Tumor suppressive protein gene associated with retinoid-interferon-induced mortality (GRIM)-19 inhibits src-induced oncogenic transformation at multiple levels
- PMID: 17823279
- PMCID: PMC1988884
- DOI: 10.2353/ajpath.2007.070241
Tumor suppressive protein gene associated with retinoid-interferon-induced mortality (GRIM)-19 inhibits src-induced oncogenic transformation at multiple levels
Abstract
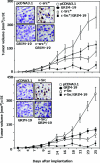
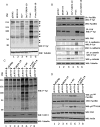
Interferons (IFNs) inhibit the growth of infectious pathogens and tumor development. Although IFNs are potent tumor suppressors, they modestly inhibit the growth of some human solid tumors. Their weak activity against such tumors is augmented by co-treatment with differentiation-inducing agents such as retinoids. Previous studies from our laboratory identified a novel gene product, gene associated with retinoid-interferon-induced mortality (GRIM)-19, as an IFN/all-trans retinoic acid-induced growth suppressor. However, the mechanisms of its growth suppressive actions are unclear. The src-family of tyrosine kinases is important regulators of various cell growth responses. Mutational activation of src causes cellular transformation by altering transcription and cytoskeletal properties. In this study, we show that GRIM-19 suppresses src-induced cellular transformation in vitro and in vivo by down-regulating the expression of a number of signal transducer and activator of transcription-3 (STAT3)-dependent cellular genes. In addition, GRIM-19 inhibited the src-induced cell motility and metastasis by suppressing the tyrosyl phosphorylation of focal adhesion kinase, paxillin, E-cadherin, and gamma-catenin. Effects of GRIM-19 on src-induced cellular transformation are reversible in the presence of specific short hairpin RNA, indicating its direct effect on transformation. GRIM-19-mediated inhibition of the src-induced tyrosyl phosphorylation of cellular proteins, such as focal adhesion kinase and paxillin, seems to occur independently of the STAT3 protein. GRIM-19 had no significant effect on the cellular transformation induced by other oncogenes such as myc and Ha-ras. Thus, GRIM-19 not only blocks src-induced gene expression through STAT3 but also the activation of cell adhesion molecules.
Figures




Similar articles
-
GRIM-19 mutations fail to inhibit v-Src-induced oncogenesis.Oncogene. 2014 Jun 12;33(24):3195-204. doi: 10.1038/onc.2013.271. Epub 2013 Jul 15. Oncogene. 2014. PMID: 23851499 Free PMC article.
-
Tumor-suppressive activity of the cell death activator GRIM-19 on a constitutively active signal transducer and activator of transcription 3.Cancer Res. 2007 Jul 1;67(13):6212-20. doi: 10.1158/0008-5472.CAN-07-0031. Cancer Res. 2007. PMID: 17616678
-
Plasmid-based Stat3-specific siRNA and GRIM-19 inhibit the growth of thyroid cancer cells in vitro and in vivo.Oncol Rep. 2014 Aug;32(2):573-80. doi: 10.3892/or.2014.3233. Epub 2014 Jun 5. Oncol Rep. 2014. PMID: 24899100
-
Cytokine-induced tumor suppressors: a GRIM story.Cytokine. 2010 Oct-Nov;52(1-2):128-42. doi: 10.1016/j.cyto.2010.03.009. Epub 2010 Apr 10. Cytokine. 2010. PMID: 20382543 Free PMC article. Review.
-
GRIM-19: A master regulator of cytokine induced tumor suppression, metastasis and energy metabolism.Cytokine Growth Factor Rev. 2017 Feb;33:1-18. doi: 10.1016/j.cytogfr.2016.09.001. Epub 2016 Sep 15. Cytokine Growth Factor Rev. 2017. PMID: 27659873 Free PMC article. Review.
Cited by
-
GRIM-19 mutations fail to inhibit v-Src-induced oncogenesis.Oncogene. 2014 Jun 12;33(24):3195-204. doi: 10.1038/onc.2013.271. Epub 2013 Jul 15. Oncogene. 2014. PMID: 23851499 Free PMC article.
-
GRIM-19 and p16(INK4a) synergistically regulate cell cycle progression and E2F1-responsive gene expression.J Biol Chem. 2010 Sep 3;285(36):27545-52. doi: 10.1074/jbc.M110.105767. Epub 2010 Jun 3. J Biol Chem. 2010. PMID: 20522552 Free PMC article.
-
The import of the transcription factor STAT3 into mitochondria depends on GRIM-19, a component of the electron transport chain.J Biol Chem. 2013 Feb 15;288(7):4723-32. doi: 10.1074/jbc.M112.378984. Epub 2012 Dec 27. J Biol Chem. 2013. PMID: 23271731 Free PMC article.
-
Identification of a structural motif in the tumor-suppressive protein GRIM-19 required for its antitumor activity.Am J Pathol. 2010 Aug;177(2):896-907. doi: 10.2353/ajpath.2010.091280. Epub 2010 Jul 1. Am J Pathol. 2010. PMID: 20595633 Free PMC article.
-
GRIM-19 inhibits v-Src-induced cell motility by interfering with cytoskeletal restructuring.Oncogene. 2009 Mar 12;28(10):1339-47. doi: 10.1038/onc.2008.480. Epub 2009 Jan 19. Oncogene. 2009. PMID: 19151760 Free PMC article.
References
-
- Kalvakolanu DV, Borden EC. Interferons: cellular and molecular biology of their actions. Bertino JR, editor. San Diego: Academic Press,; Encyclopedia of Cancer. 2002:pp 511–521.
-
- Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006;6:836–848. - PubMed
-
- Kalvakolanu DV. The GRIMs: a new interface between cell death regulation and interferon/retinoid induced growth suppression. Cytokine Growth Factor Rev. 2004;15:169–194. - PubMed
-
- Lindner DJ, Borden EC, Kalvakolanu DV. Synergistic antitumor effects of a combination of interferons and retinoic acid on human tumor cells in vitro and in vivo. Clin Cancer Res. 1997;3:931–937. - PubMed
-
- Lippman SM, Kalvakolanu DV, Lotan R. Retinoids and interferons in non-melanoma skin cancer. J Investig Dermatol Symp Proc. 1996;1:219–222. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA105005/CA/NCI NIH HHS/United States
- HL084223/HL/NHLBI NIH HHS/United States
- ES11863/ES/NIEHS NIH HHS/United States
- R01 CA105005/CA/NCI NIH HHS/United States
- R01 HL084223/HL/NHLBI NIH HHS/United States
- CA78282/CA/NCI NIH HHS/United States
- HL66109/HL/NHLBI NIH HHS/United States
- R01 HL066109/HL/NHLBI NIH HHS/United States
- R56 ES011863/ES/NIEHS NIH HHS/United States
- R01 ES011863/ES/NIEHS NIH HHS/United States
- CA095020/CA/NCI NIH HHS/United States
- R01 CA078282/CA/NCI NIH HHS/United States
- R01 CA095020/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

