Dual localization pathways for the engulfment proteins during Bacillus subtilis sporulation
- PMID: 17824930
- PMCID: PMC2885130
- DOI: 10.1111/j.1365-2958.2007.05887.x
Dual localization pathways for the engulfment proteins during Bacillus subtilis sporulation
Abstract
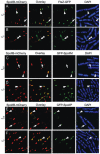
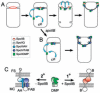
Engulfment in Bacillus subtilis is mediated by two complementary systems, SpoIID, SpoIIM and SpoIIP (DMP), which are essential for engulfment, and the SpoIIQ-SpoIIIAGH (Q-AH) zipper, which provides a secondary engulfment mechanism and recruits other proteins to the septum. We here identify two mechanisms by which DMP localizes to the septum. The first depends on SpoIIB, which is recruited to the septum during division and provides a septal landmark for efficient DMP localization. However, sporangia lacking SpoIIB ultimately localize DMP and complete engulfment, suggesting a second mechanism for DMP localization. This secondary targeting pathway depends on SpoIVFA and SpoIVFB, which are recruited to the septum by the Q-AH zipper. The absence of a detectable localization phenotype in mutants lacking only SpoIVFAB (or Q-AH) suggests that SpoIIB provides the primary DMP localization pathway while SpoIVFAB provides a secondary pathway. In keeping with this hypothesis, the spoIIB spoIVFAB mutant strain has a synergistic engulfment defect at septal thinning (which requires DMP) and is completely defective in DMP localization. Thus, the Q-AH zipper both provides a compensatory mechanism for engulfment when DMP activity is reduced, and indirectly provides a compensatory mechanism for septal localization of DMP when its primary targeting pathway is disrupted.
Figures

References
-
- Chastanet A, Losick R. Engulfment during sporulation in Bacillus subtilis is governed by a multi-protein complex containing tandemly acting autolysins. Mol Microbiol. 2007;64:139–152. - PubMed
-
- Chen JC, Viollier PH, Shapiro L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol Microbiol. 2005;55:1085–1103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

