An ATPase activity associated with the rotavirus phosphoprotein NSP5
- PMID: 17825341
- PMCID: PMC2702534
- DOI: 10.1016/j.virol.2007.07.029
An ATPase activity associated with the rotavirus phosphoprotein NSP5
Abstract
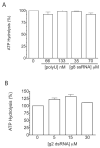
Interactions between NSP5 and NSP2 drive the formation of viroplasms, sites of genome replication and packaging in rotavirus-infected cells. The serine-threonine-rich NSP5 transitions between hypo- and hyper-phosphorylated isomers during the replication cycle. In this study, we determined that purified recombinant NSP5 has a Mg2+-dependent ATP-specific triphosphatase activity that generates free ADP and Pi (Vmax of 19.33 fmol of product/min/pmol of enzyme). The ATPase activity was correlated with low levels of NSP5 phosphorylation, suggestive of a possible link between ATP hydrolysis and an NSP5 autokinase activity. Mutagenesis showed that the critical residue (Ser67) needed for NSP5 hyperphosphorylation by cellular casein kinase-like enzymes has no role in the ATPase or autokinase activities of NSP5. Through its NDP kinase activity, the NSP2 octamer may support NSP5 phosphorylation by creating a constant source of ATP molecules for the autokinase activity of NSP5 and for cellular kinases associated with NSP5.
Figures
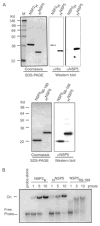
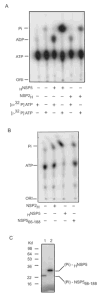
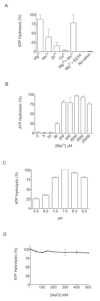

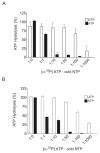



References
-
- Afrikanova I, Fabbretti E, Miozzo M, Burrone O. Rotavirus NSP5 phosphorylation is up-regulated by interaction with NSP2. J Gen Virol. 1998;79:2679–2686. - PubMed
-
- Afrikanova I, Miozzo M, Giambiagi S, Burrone O. Phosphorylation generates different forms of rotavirus NSP5. J Gen Virol. 1996;77:2059–2065. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

