Pseudoknot structures with conserved base triples in telomerase RNAs of ciliates
- PMID: 17827211
- PMCID: PMC2094054
- DOI: 10.1093/nar/gkm660
Pseudoknot structures with conserved base triples in telomerase RNAs of ciliates
Abstract
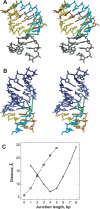
Telomerase maintains the integrity of telomeres, the ends of linear chromosomes, by adding G-rich repeats to their 3'-ends. Telomerase RNA is an integral component of telomerase. It contains a template for the synthesis of the telomeric repeats by the telomerase reverse transcriptase. Although telomerase RNAs of different organisms are very diverse in their sequences, a functional non-template element, a pseudoknot, was predicted in all of them. Pseudoknot elements in human and the budding yeast Kluyveromyces lactis telomerase RNAs contain unusual triple-helical segments with AUU base triples, which are critical for telomerase function. Such base triples in ciliates have not been previously reported. We analyzed the pseudoknot sequences in 28 ciliate species and classified them in six different groups based on the lengths of the stems and loops composing the pseudoknot. Using miniCarlo, a helical parameter-based modeling program, we calculated 3D models for a representative of each morphological group. In all cases, the predicted structure contains at least one AUU base triple in stem 2, except for that of Colpidium colpoda, which contains unconventional GCG and AUA triples. These results suggest that base triples in a pseudoknot element are a conserved feature of all telomerases.
Figures

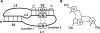

References
-
- Bertuch AA, Lundblad V. The maintenance and masking of chromosome termini. Curr. Opin. Cell Biol. 2006;18:247–253. - PubMed
-
- Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 2006;75:493–517. - PubMed
-
- Romero DP, Blackburn EH. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. - PubMed
-
- Lingner J, Hendrick LL, Cech TR. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev. 1994;8:1984–1998. - PubMed
-
- Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. - PubMed

