Splicing-independent recruitment of spliceosomal small nuclear RNPs to nascent RNA polymerase II transcripts
- PMID: 17846169
- PMCID: PMC2064619
- DOI: 10.1083/jcb.200706134
Splicing-independent recruitment of spliceosomal small nuclear RNPs to nascent RNA polymerase II transcripts
Abstract
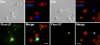
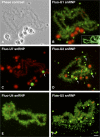
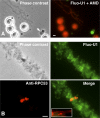
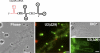
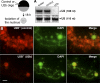
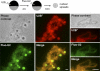
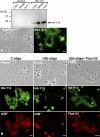
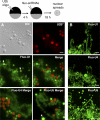
In amphibian oocytes, most lateral loops of the lampbrush chromosomes correspond to active transcriptional sites for RNA polymerase II. We show that newly assembled small nuclear ribonucleoprotein (RNP [snRNP]) particles, which are formed upon cytoplasmic injection of fluorescently labeled spliceosomal small nuclear RNAs (snRNAs), target the nascent transcripts of the chromosomal loops. With this new targeting assay, we demonstrate that nonfunctional forms of U1 and U2 snRNAs still associate with the active transcriptional units. In particular, we find that their association with nascent RNP fibrils is independent of their base pairing with pre-messenger RNAs. Additionally, stem loop I of the U1 snRNA is identified as a discrete domain that is both necessary and sufficient for association with nascent transcripts. Finally, in oocytes deficient in splicing, the recruitment of U1, U4, and U5 snRNPs to transcriptional units is not affected. Collectively, these data indicate that the recruitment of snRNPs to nascent transcripts and the assembly of the spliceosome are uncoupled events.
Figures


References
-
- Aguilera, A. 2005. Cotranscriptional mRNP assembly: from the DNA to the nuclear pore. Curr. Opin. Cell Biol. 17:242–250. - PubMed
-
- Beenders, B., E. Watrin, V. Legagneux, I. Kireev, and M. Bellini. 2003. Distribution of XCAP-E and XCAP-D2 in the Xenopus oocyte nucleus. Chromosome Res. 11:549–564. - PubMed
-
- Bentley, D.L. 2005. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 17:251–256. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

