TBX22 missense mutations found in patients with X-linked cleft palate affect DNA binding, sumoylation, and transcriptional repression
- PMID: 17846996
- PMCID: PMC2227921
- DOI: 10.1086/521033
TBX22 missense mutations found in patients with X-linked cleft palate affect DNA binding, sumoylation, and transcriptional repression
Abstract
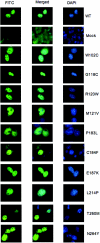
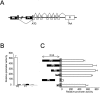
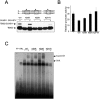
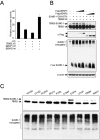
The T-box transcription factor TBX22 is essential for normal craniofacial development, as demonstrated by the finding of nonsense, frameshift, splice-site, or missense mutations in patients with X-linked cleft palate (CPX) and ankyloglossia. To better understand the function of TBX22, we studied 10 different naturally occurring missense mutations that are phenotypically equivalent to loss-of-function alleles. Since all missense mutations are located in the DNA-binding T-box domain, we first investigated the preferred recognition sequence for TBX22. Typical of T-box proteins, the resulting sequence is a palindrome based around near-perfect copies of AGGTGTGA. DNA-binding assays indicate that missense mutations at or near predicted contact points with the DNA backbone compromise stable DNA-protein interactions. We show that TBX22 functions as a transcriptional repressor and that TBX22 missense mutations result in impaired repression activity. No effect on nuclear localization of TBX22 was observed. We find that TBX22 is a target for the small ubiquitin-like modifier SUMO-1 and that this modification is required for TBX22 repressor activity. Although the site of SUMO attachment at the lysine at position 63 is upstream of the T-box domain, loss of SUMO-1 modification is consistently found in all pathogenic CPX missense mutations. This implies a general mechanism linking the loss of SUMO conjugation to the loss of TBX22 function. Orofacial clefts are well known for their complex etiology and variable penetrance, involving both genetic and environmental risk factors. The sumoylation process is also subject to and profoundly affected by similar environmental stresses. Thus, we suggest that SUMO modification may represent a common pathway that regulates normal craniofacial development and is involved in the pathogenesis of both Mendelian and idiopathic forms of orofacial clefting.
Figures




References
Web Resources
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CPX) - PubMed
-
- UCSC Genome Browser, http://genome.ucsc.edu/
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

