A Bayesian approach to copy-number-polymorphism analysis in nuclear pedigrees
- PMID: 17847005
- PMCID: PMC2227930
- DOI: 10.1086/520096
A Bayesian approach to copy-number-polymorphism analysis in nuclear pedigrees
Abstract
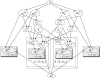
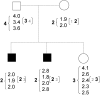
Segmental copy-number polymorphisms (CNPs) represent a significant component of human genetic variation and are likely to contribute to disease susceptibility. These potentially multiallelic and highly polymorphic systems present new challenges to family-based genetic-analysis tools that commonly assume codominant markers and allow for no genotyping error. The copy-number quantitation (CNP phenotype) represents the total number of segmental copies present in an individual and provides a means to infer, rather than to observe, the underlying allele segregation. We present an integrated approach to meet these challenges, in the form of a graphical model in which we infer the underlying CNP phenotype from the (single or replicate) quantitative measure within the analysis while assuming an allele-based system segregating through the pedigree. This approach can be readily applied to the study of any form of genetic measure, and the construction permits extension to a wide variety of hypothesis tests. We have implemented the basic model for use with nuclear families, and we illustrate its application through an analysis of the CNP located in gene CCL3L1 in 201 families with asthma.
Figures
Similar articles
-
Partitioning of copy-number genotypes in pedigrees.BMC Bioinformatics. 2010 May 3;11:226. doi: 10.1186/1471-2105-11-226. BMC Bioinformatics. 2010. PMID: 20438641 Free PMC article.
-
Multiplex Paralogue Ratio Tests for accurate measurement of multiallelic CNVs.Genomics. 2009 Jan;93(1):98-103. doi: 10.1016/j.ygeno.2008.09.004. Epub 2008 Oct 22. Genomics. 2009. PMID: 18848619
-
PlatinumCNV: a Bayesian Gaussian mixture model for genotyping copy number polymorphisms using SNP array signal intensity data.Genet Epidemiol. 2011 Dec;35(8):831-44. doi: 10.1002/gepi.20633. Genet Epidemiol. 2011. PMID: 22125222
-
Copy-number variation and association studies of human disease.Nat Genet. 2007 Jul;39(7 Suppl):S37-42. doi: 10.1038/ng2080. Nat Genet. 2007. PMID: 17597780 Review.
-
Copy-number polymorphisms: mining the tip of an iceberg.Trends Genet. 2005 Jun;21(6):315-7. doi: 10.1016/j.tig.2005.04.007. Trends Genet. 2005. PMID: 15922827 Review.
Cited by
-
Modeling genetic inheritance of copy number variations.Nucleic Acids Res. 2008 Dec;36(21):e138. doi: 10.1093/nar/gkn641. Epub 2008 Oct 2. Nucleic Acids Res. 2008. PMID: 18832372 Free PMC article.
-
Reconstructing CNV genotypes using segregation analysis: combining pedigree information with CNV assay.Genet Sel Evol. 2010 Aug 12;42(1):34. doi: 10.1186/1297-9686-42-34. Genet Sel Evol. 2010. PMID: 20701809 Free PMC article.
-
Detection, imputation, and association analysis of small deletions and null alleles on oligonucleotide arrays.Am J Hum Genet. 2008 Jun;82(6):1316-33. doi: 10.1016/j.ajhg.2008.05.008. Am J Hum Genet. 2008. PMID: 18519066 Free PMC article.
-
Genetic association analysis of copy-number variation (CNV) in human disease pathogenesis.Genomics. 2009 Jan;93(1):22-6. doi: 10.1016/j.ygeno.2008.08.012. Epub 2008 Oct 19. Genomics. 2009. PMID: 18822366 Free PMC article. Review.
-
A stochastic inference of de novo CNV detection and association test in multiplex schizophrenia families.Front Genet. 2013 Sep 23;4:185. doi: 10.3389/fgene.2013.00185. eCollection 2013. Front Genet. 2013. PMID: 24065985 Free PMC article.
References
Web Resources
-
- Database of Genomic Variants, http://projects.tcag.ca/variation/
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

