Solution structure of the second bromodomain of Brd2 and its specific interaction with acetylated histone tails
- PMID: 17848202
- PMCID: PMC2065866
- DOI: 10.1186/1472-6807-7-57
Solution structure of the second bromodomain of Brd2 and its specific interaction with acetylated histone tails
Abstract
Background: Brd2 is a transcriptional regulator and belongs to BET family, a less characterized novel class of bromodomain-containing proteins. Brd2 contains two tandem bromodomains (BD1 and BD2, 46% sequence identity) in the N-terminus and a conserved motif named ET (extra C-terminal) domain at the C-terminus that is also present in some other bromodomain proteins. The two bromodomains have been shown to bind the acetylated histone H4 and to be responsible for mitotic retention on chromosomes, which is probably a distinctive feature of BET family proteins. Although the crystal structure of Brd2 BD1 is reported, no structure features have been characterized for Brd2 BD2 and its interaction with acetylated histones.
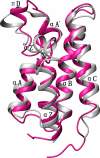
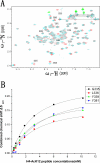
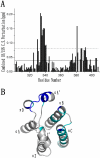
Results: Here we report the solution structure of human Brd2 BD2 determined by NMR. Although the overall fold resembles the bromodomains from other proteins, significant differences can be found in loop regions, especially in the ZA loop in which a two amino acids insertion is involved in an uncommon pi-helix, termed piD. The helix piD forms a portion of the acetyl-lysine binding site, which could be a structural characteristic of Brd2 BD2 and other BET bromodomains. Unlike Brd2 BD1, BD2 is monomeric in solution. With NMR perturbation studies, we have mapped the H4-AcK12 peptide binding interface on Brd2 BD2 and shown that the binding was with low affinity (2.9 mM) and in fast exchange. Using NMR and mutational analysis, we identified several residues important for the Brd2 BD2-H4-AcK12 peptide interaction and probed the potential mechanism for the specific recognition of acetylated histone codes by Brd2 BD2.
Conclusion: Brd2 BD2 is monomeric in solution and dynamically interacts with H4-AcK12. The additional secondary elements in the long ZA loop may be a common characteristic of BET bromodomains. Surrounding the ligand-binding cavity, five aspartate residues form a negatively charged collar that serves as a secondary binding site for H4-AcK12. We suggest that Brd2 BD1 and BD2 may possess distinctive roles and cooperate to regulate Brd2 functions. The structure basis of Brd2 BD2 will help to further characterize the functions of Brd2 and its BET members.
Figures
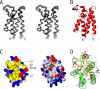


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

