Conceptual modeling in systems biology fosters empirical findings: the mRNA lifecycle
- PMID: 17849002
- PMCID: PMC1964809
- DOI: 10.1371/journal.pone.0000872
Conceptual modeling in systems biology fosters empirical findings: the mRNA lifecycle
Abstract
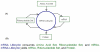
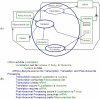
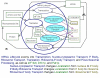
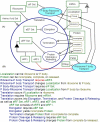
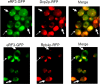
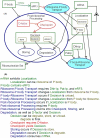
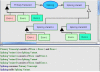
One of the main obstacles to understanding complex biological systems is the extent and rapid evolution of information, way beyond the capacity individuals to manage and comprehend. Current modeling approaches and tools lack adequate capacity to model concurrently structure and behavior of biological systems. Here we propose Object-Process Methodology (OPM), a holistic conceptual modeling paradigm, as a means to model both diagrammatically and textually biological systems formally and intuitively at any desired number of levels of detail. OPM combines objects, e.g., proteins, and processes, e.g., transcription, in a way that is simple and easily comprehensible to researchers and scholars. As a case in point, we modeled the yeast mRNA lifecycle. The mRNA lifecycle involves mRNA synthesis in the nucleus, mRNA transport to the cytoplasm, and its subsequent translation and degradation therein. Recent studies have identified specific cytoplasmic foci, termed processing bodies that contain large complexes of mRNAs and decay factors. Our OPM model of this cellular subsystem, presented here, led to the discovery of a new constituent of these complexes, the translation termination factor eRF3. Association of eRF3 with processing bodies is observed after a long-term starvation period. We suggest that OPM can eventually serve as a comprehensive evolvable model of the entire living cell system. The model would serve as a research and communication platform, highlighting unknown and uncertain aspects that can be addressed empirically and updated consequently while maintaining consistency.
Conflict of interest statement
Figures
References
-
- Kitano H. Systems Biology: A Brief Overview. Science, 2002;295(5560):1662–1664. - PubMed
-
- Kritikou E, Pulverer B, Heinrichs A. Editorial to “Systems Biology: A User's Guide”. Joint online resource by Nature Reviews Molecular Cell Biology and Nature Cell Biology, 2006. http://www.nature.com/focus/systemsbiologyuserguide/editorial/sysbiol-s3....
-
- Davidson EH, et al. A Genomic Regulatory Network for Development. Science, 2002;295(5560):1669–1678. - PubMed
-
- Tomita M. Whole-cell simulation: a grand challenge of the 21st century. Trends Biotechnol, 2001;19(6):205–10. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

