Activation of the Cdc42p GTPase by cyclin-dependent protein kinases in budding yeast
- PMID: 17853895
- PMCID: PMC2063489
- DOI: 10.1038/sj.emboj.7601847
Activation of the Cdc42p GTPase by cyclin-dependent protein kinases in budding yeast
Abstract
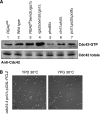
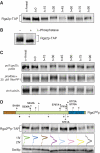
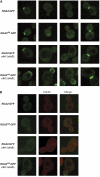
Cyclin-dependent kinases (CDKs) trigger essential cell cycle processes including critical events in G1 phase that culminate in bud emergence, spindle pole body duplication, and DNA replication. Localized activation of the Rho-type GTPase Cdc42p is crucial for establishment of cell polarity during G1, but CDK targets that link the Cdc42p module with cell growth and cell cycle commitment have remained largely elusive. Here, we identify the GTPase-activating protein (GAP) Rga2p as an important substrate related to the cell polarity function of G1 CDKs. Overexpression of RGA2 in the absence of functional Pho85p or Cdc28p CDK complexes is toxic, due to an inability to polarize growth. Mutation of CDK consensus sites in Rga2p that are phosphorylated both in vivo and in vitro by Pho85p and Cdc28p CDKs results in a loss of G1 phase-specific phosphorylation. A failure to phosphorylate Rga2p leads to defects in localization and impaired polarized growth, in a manner dependent on Rga2p GAP function. Taken together, our data suggest that CDK-dependent phosphorylation restrains Rga2p activity to ensure appropriate activation of Cdc42p during cell polarity establishment. Inhibition of GAPs by CDK phosphorylation may be a general mechanism to promote proper G1-phase progression.
Figures
References
-
- Archambault V, Chang EJ, Drapkin BJ, Cross FR, Chait BT, Rout MP (2004) Targeted proteomic study of the cyclin–Cdk module. Mol Cell 14: 699–711 - PubMed
-
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM (2000) A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407: 395–401 - PubMed
-
- Bloom J, Cross FR (2007) Multiple levels of cyclin specificity in cell-cycle control. Nat Rev Mol Cell Biol 8: 149–160 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

