The role of iron in Mycobacterium smegmatis biofilm formation: the exochelin siderophore is essential in limiting iron conditions for biofilm formation but not for planktonic growth
- PMID: 17854402
- PMCID: PMC2170428
- DOI: 10.1111/j.1365-2958.2007.05935.x
The role of iron in Mycobacterium smegmatis biofilm formation: the exochelin siderophore is essential in limiting iron conditions for biofilm formation but not for planktonic growth
Abstract
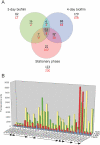
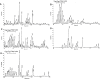
Many species of mycobacteria form structured biofilm communities at liquid-air interfaces and on solid surfaces. Full development of Mycobacterium smegmatis biofilms requires addition of supplemental iron above 1 microM ferrous sulphate, although addition of iron is not needed for planktonic growth. Microarray analysis of the M. smegmatis transcriptome shows that iron-responsive genes - especially those involved in siderophore synthesis and iron uptake - are strongly induced during biofilm formation reflecting a response to iron deprivation, even when 2 microM iron is present. The acquisition of iron under these conditions is specifically dependent on the exochelin synthesis and uptake pathways, and the strong defect of an iron-exochelin uptake mutant suggests a regulatory role of iron in the transition to biofilm growth. In contrast, although the expression of mycobactin and iron ABC transport operons is highly upregulated during biofilm formation, mutants in these systems form normal biofilms in low-iron (2 microM) conditions. A close correlation between iron availability and matrix-associated fatty acids implies a possible metabolic role in the late stages of biofilm maturation, in addition to the early regulatory role. M. smegmatis surface motility is similarly dependent on iron availability, requiring both supplemental iron and the exochelin pathway to acquire it.
Figures






Similar articles
-
Identification of genes involved in the sequestration of iron in mycobacteria: the ferric exochelin biosynthetic and uptake pathways.Mol Microbiol. 1994 Nov;14(3):557-69. doi: 10.1111/j.1365-2958.1994.tb02189.x. Mol Microbiol. 1994. PMID: 7885234
-
Exochelin genes in Mycobacterium smegmatis: identification of an ABC transporter and two non-ribosomal peptide synthetase genes.Mol Microbiol. 1998 Jul;29(2):629-39. doi: 10.1046/j.1365-2958.1998.00961.x. Mol Microbiol. 1998. PMID: 9720878
-
Role of a 21-kDa iron-regulated protein IrpA in the uptake of ferri-exochelin by Mycobacterium smegmatis.J Appl Microbiol. 2020 Dec;129(6):1733-1743. doi: 10.1111/jam.14728. Epub 2020 Jun 25. J Appl Microbiol. 2020. PMID: 32472729
-
Proteomics and mass spectrometric studies reveal planktonic growth of Mycobacterium smegmatis in biofilm cultures in the absence of rpoZ.J Chromatogr B Analyt Technol Biomed Life Sci. 2008 Jan 15;861(2):196-202. doi: 10.1016/j.jchromb.2007.08.009. Epub 2007 Aug 21. J Chromatogr B Analyt Technol Biomed Life Sci. 2008. PMID: 17822967
-
Lessons from DNA microarray analysis: the gene expression profile of biofilms.Curr Opin Microbiol. 2005 Apr;8(2):222-7. doi: 10.1016/j.mib.2005.02.015. Curr Opin Microbiol. 2005. PMID: 15802256 Review.
Cited by
-
Targeting drug tolerance in mycobacteria: a perspective from mycobacterial biofilms.Expert Rev Anti Infect Ther. 2012 Sep;10(9):1055-66. doi: 10.1586/eri.12.88. Expert Rev Anti Infect Ther. 2012. PMID: 23106280 Free PMC article. Review.
-
Functions of the periplasmic loop of the porin MspA from Mycobacterium smegmatis.J Biol Chem. 2009 Apr 10;284(15):10223-31. doi: 10.1074/jbc.M808599200. Epub 2009 Feb 10. J Biol Chem. 2009. PMID: 19208627 Free PMC article.
-
Production of mycobacterial cell wall glycopeptidolipids requires a member of the MbtH-like protein family.BMC Microbiol. 2012 Jun 22;12:118. doi: 10.1186/1471-2180-12-118. BMC Microbiol. 2012. PMID: 22726990 Free PMC article.
-
Conversion of carbon dioxide in biogas into acetic acid by Clostridium thailandense immobilized on porous support materials.Heliyon. 2024 Feb 13;10(4):e26378. doi: 10.1016/j.heliyon.2024.e26378. eCollection 2024 Feb 29. Heliyon. 2024. PMID: 38390190 Free PMC article.
-
The extracellular matrix of mycobacterial biofilms: could we shorten the treatment of mycobacterial infections?Microb Cell. 2019 Jan 18;6(2):105-122. doi: 10.15698/mic2019.02.667. Microb Cell. 2019. PMID: 30740456 Free PMC article. Review.
References
-
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. - PubMed
-
- Bardarov S, Bardarov S, Pavelka MS, Sambandamurthy V, Larsen M, Tufariello J, et al. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology. 2002;148:3007–3017. - PubMed
-
- Beloin C, Ghigo JM. Finding gene-expression patterns in bacterial biofilms. Trends Microbiol. 2005;13:16–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

