DNA evidence for global dispersal and probable endemicity of protozoa
- PMID: 17854485
- PMCID: PMC2194784
- DOI: 10.1186/1471-2148-7-162
DNA evidence for global dispersal and probable endemicity of protozoa
Abstract
Background: It is much debated whether microbes are easily dispersed globally or whether they, like many macro-organisms, have historical biogeographies. The ubiquitous dispersal hypothesis states that microbes are so numerous and so easily dispersed worldwide that all should be globally distributed and found wherever growing conditions suit them. This has been broadly upheld for protists (microbial eukaryotes) by most morphological and some molecular analyses. However, morphology and most previously used evolutionary markers evolve too slowly to test this important hypothesis adequately.
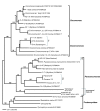
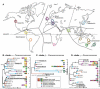
Results: Here we use a fast-evolving marker (ITS1 rDNA) to map global diversity and distribution of three different clades of cercomonad Protozoa (Eocercomonas and Paracercomonas: phylum Cercozoa) by sequencing multiple environmental gene libraries constructed from 47-80 globally-dispersed samples per group. Even with this enhanced resolution, identical ITS sequences (ITS-types) were retrieved from widely separated sites and on all continents for several genotypes, implying relatively rapid global dispersal. Some identical ITS-types were even recovered from both marine and non-marine samples, habitats that generally harbour significantly different protist communities. Conversely, other ITS-types had either patchy or restricted distributions.
Conclusion: Our results strongly suggest that geographic dispersal in macro-organisms and microbes is not fundamentally different: some taxa show restricted and/or patchy distributions while others are clearly cosmopolitan. These results are concordant with the 'moderate endemicity model' of microbial biogeography. Rare or continentally endemic microbes may be ecologically significant and potentially of conservational concern. We also demonstrate that strains with identical 18S but different ITS1 rDNA sequences can differ significantly in terms of morphological and important physiological characteristics, providing strong additional support for global protist biodiversity being significantly higher than previously thought.
Figures


References
-
- Baas-Becking LGM. Geobiologie of Inleiding tot de Milieukunde. In: van Stockum WP, Zoon NV, editor. The Hague. The Netherlands ; 1934.
-
- Hughes Martiny JB, Bohannan BJM, Brown JH, Colwell RK, Fuhrmann JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Øvreås L, Reysenback AL, Smith VH, Staley JT. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. - DOI - PubMed
-
- Finlay BJ, Fenchel T. Divergent perspectives on protist species richness. Protist. 1999;150:229–233. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

