Molecular evolution of type VI intermediate filament proteins
- PMID: 17854500
- PMCID: PMC2075511
- DOI: 10.1186/1471-2148-7-164
Molecular evolution of type VI intermediate filament proteins
Abstract
Background: Tanabin, transitin and nestin are type VI intermediate filament (IF) proteins that are developmentally regulated in frogs, birds and mammals, respectively. Tanabin is expressed in the growth cones of embryonic vertebrate neurons, whereas transitin and nestin are found in myogenic and neurogenic cells. Another type VI IF protein, synemin, is expressed in undifferentiated and mature muscle cells of birds and mammals. In addition to an IF-typical alpha-helical core domain, type VI IF proteins are characterized by a long C-terminal tail often containing distinct repeated motifs. The molecular evolution of type VI IF proteins remains poorly studied.
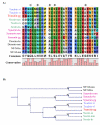
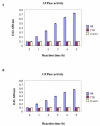
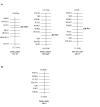
Results: To examine the evolutionary history of type VI IF proteins, sequence comparisons, BLAST searches, synteny studies and phylogenic analyses were performed. This study provides new evidence that tanabin, transitin and nestin are indeed orthologous type VI IF proteins. It demonstrates that tanabin, transitin and nestin genes share intron positions and sequence identities, have a similar chromosomal context and display closely related positions in phylogenic analyses. Despite this homology, fast evolution rates of their C-terminal extremity have caused the appearance of repeated motifs with distinct biological activities. In particular, our in silico and in vitro analyses of their tail domain have shown that (avian) transitin, but not (mammalian) nestin, contains a repeat domain displaying nucleotide hydrolysis activity.
Conclusion: These analyses of the evolutionary history of the IF proteins fit with a model in which type VI IFs form a branch distinct from NF proteins and are composed of two major proteins: synemin and nestin orthologs. Rapid evolution of the C-terminal extremity of nestin orthologs could be responsible for their divergent functions.
Figures


References
-
- Hesse M, Magin TM, Weber K. Genes for intermediate filament proteins and the draft sequence of the human genome: novel keratin genes and a surprisingly high number of pseudogenes related to keratin genes 8 and 18. J Cell Sci. 2001;114:2569–2575. - PubMed
-
- Parry DA. Microdissection of the sequence and structure of intermediate filament chains. Adv Protein Chem. 2005;70:113–142. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

