DNA sequencing: bench to bedside and beyond
- PMID: 17855400
- PMCID: PMC2094077
- DOI: 10.1093/nar/gkm688
DNA sequencing: bench to bedside and beyond
Abstract
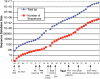
Fifteen years elapsed between the discovery of the double helix (1953) and the first DNA sequencing (1968). Modern DNA sequencing began in 1977, with development of the chemical method of Maxam and Gilbert and the dideoxy method of Sanger, Nicklen and Coulson, and with the first complete DNA sequence (phage X174), which demonstrated that sequence could give profound insights into genetic organization. Incremental improvements allowed sequencing of molecules >200 kb (human cytomegalovirus) leading to an avalanche of data that demanded computational analysis and spawned the field of bioinformatics. The US Human Genome Project spurred sequencing activity. By 1992 the first 'sequencing factory' was established, and others soon followed. The first complete cellular genome sequences, from bacteria, appeared in 1995 and other eubacterial, archaebacterial and eukaryotic genomes were soon sequenced. Competition between the public Human Genome Project and Celera Genomics produced working drafts of the human genome sequence, published in 2001, but refinement and analysis of the human genome sequence will continue for the foreseeable future. New 'massively parallel' sequencing methods are greatly increasing sequencing capacity, but further innovations are needed to achieve the 'thousand dollar genome' that many feel is prerequisite to personalized genomic medicine. These advances will also allow new approaches to a variety of problems in biology, evolution and the environment.
Figures
References
-
- Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977;265:687–695. - PubMed
-
- Sanger F, Coulson AR, Friedmann T, Air GM, Barrell BG, Brown NL, Fiddes JC, Hutchison CA, III, Slocombe PM, et al. The nucleotide sequence of bacteriophage phiX174. J. Mol. Biol. 1978;125:225–246. - PubMed
-
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

