Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication
- PMID: 17855519
- PMCID: PMC2168994
- DOI: 10.1128/JVI.01506-07
Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication
Abstract
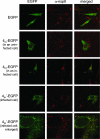
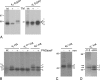
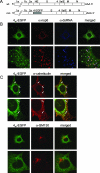
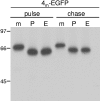
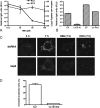
The coronavirus nonstructural proteins (nsp's) derived from the replicase polyproteins collectively constitute the viral replication complexes, which are anchored to double-membrane vesicles. Little is known about the biogenesis of these complexes, the membrane anchoring of which is probably mediated by nsp3, nsp4, and nsp6, as they contain several putative transmembrane domains. As a first step to getting more insight into the formation of the coronavirus replication complex, the membrane topology, processing, and subcellular localization of nsp4 of the mouse hepatitis virus (MHV) and severe acute respiratory syndrome-associated coronavirus (SARS-CoV) were elucidated in this study. Both nsp4 proteins became N glycosylated, while their amino and carboxy termini were localized to the cytoplasm. These observations imply nsp4 to assemble in the membrane as a tetraspanning transmembrane protein with a Nendo/Cendo topology. The amino terminus of SARS-CoV nsp4, but not that of MHV nsp4, was shown to be (partially) processed by signal peptidase. nsp4 localized to the endoplasmic reticulum (ER) when expressed alone but was recruited to the replication complexes in infected cells. nsp4 present in these complexes did not colocalize with markers of the ER or Golgi apparatus, while the susceptibility of its sugars to endoglycosidase H indicated that the protein had also not traveled trough the latter compartment. The important role of the early secretory pathway in formation of the replication complexes was also demonstrated by the inhibition of coronaviral replication when the ER export machinery was blocked by use of the kinase inhibitor H89 or by expression of a mutant, Sar1[H79G].
Figures
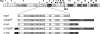


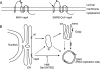
References
-
- Altan-Bonnet, N., R. Sougrat, and J. Lippincott-Schwartz. 2004. Molecular basis for Golgi maintenance and biogenesis. Curr. Opin. Cell Biol. 16:364-372. - PubMed
-
- Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. - PubMed
-
- Aridor, M., and W. E. Balch. 2000. Kinase signaling initiates coat complex II (COPII) recruitment and export from the mammalian endoplasmic reticulum. J. Biol. Chem. 275:35673-35676. - PubMed
-
- Belov, G. A., and E. Ehrenfeld. 2007. Involvement of cellular membrane traffic proteins in poliovirus replication. Cell Cycle 6:36-38. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

