Molecular characterization of human immunodeficiency virus type 1 (HIV-1) and HIV-2 in Yaounde, Cameroon: evidence of major drug resistance mutations in newly diagnosed patients infected with subtypes other than subtype B
- PMID: 17855574
- PMCID: PMC2224252
- DOI: 10.1128/JCM.00428-07
Molecular characterization of human immunodeficiency virus type 1 (HIV-1) and HIV-2 in Yaounde, Cameroon: evidence of major drug resistance mutations in newly diagnosed patients infected with subtypes other than subtype B
Abstract
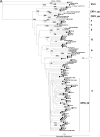
Prior to current studies on the emergence of drug resistance with the introduction of antiretroviral therapy (ART) in Cameroon, we performed genotypic analysis on samples from drug-naïve, human immunodeficiency virus (HIV)-infected individuals in this country. Of the 79 HIV type 1 (HIV-1) pol sequences analyzed from Cameroonian samples, 3 (3.8%) were identified as HIV-1 group O, 1 (1.2%) was identified as an HIV-2 intergroup B/A recombinant, and the remaining 75 (95.0%) were identified as HIV-1 group M. Group M isolates were further classified as subtypes A1 (n = 4), D (n = 4), F2 (n = 6), G (n = 12), H (n = 2), and K (n = 1) and as circulating recombinant forms CRF02_AG (n = 41), CRF11_cpx (n = 1), and CRF13_cpx (n = 2). Two pol sequences were identified as unique recombinant forms of CRF02_AG/F2 (n = 2). M46L (n = 2), a major resistance mutation associated with resistance to protease inhibitors, was observed in 2/75 (2.6%) group M samples. Single mutations associated with resistance to nucleoside reverse transcriptase inhibitors (T215Y/F [n = 3]) and nonnucleoside reverse transcriptase inhibitors (V108I [n = 1], L100I [n = 1], and Y181C [n = 2]) were observed in 7 of 75 (9.3%) group M samples. None of the patients had any history of ART exposure. Population surveillance of transmitted HIV drug resistance is required and should be included to aid in the development of appropriate guidelines.
Figures


References
-
- Aghokeng, A. F., L. Ewane, B. Awazi, A. Nanfack, E. Delaporte, L. Zekeng, and M. Peeters. 2005. Enfuvirtide binding domain is highly conserved in non-B HIV type 1 strains from Cameroon, West Central Africa. AIDS Res. Hum. Retrovir. 21430-433. - PubMed
-
- Brenner, B., D. Turner, M. Oliveira, D. Moisi, M. Detorio, M. Carobene, R. G. Marlink, J. Schapiro, M. Roger, and M. A. Wainberg. 2003. A V106 mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non nucleoside reverse transcriptase inhibitors. AIDS 17F1-F5. - PubMed
-
- Bussmann, H., V. Novitsky, and W. Wester. 2002. Frequency of drug resistant mutations among HIV-1 C infected drug-naïve patients in Botswana, abstr. TuPeB4607. Abstr. 14th Int. AIDS Conf., Barcelona, Spain.
-
- Descamps, D., G. Collin, F. Letourneur, C. Apetrei, F. Damon, I. Loussert-Ajaka, F. Simon, S. Saragosti, and F. Brun-Vezinet. 1997. Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: in vitro phenotypic and genotypic analyses. J. Virol. 718893-8898. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

