A single injection of an adeno-associated virus vector into nuclei with divergent connections results in widespread vector distribution in the brain and global correction of a neurogenetic disease
- PMID: 17855607
- PMCID: PMC6672652
- DOI: 10.1523/JNEUROSCI.2185-07.2007
A single injection of an adeno-associated virus vector into nuclei with divergent connections results in widespread vector distribution in the brain and global correction of a neurogenetic disease
Abstract
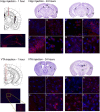
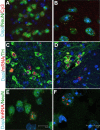
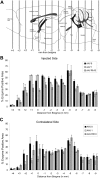
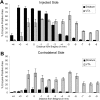
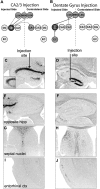
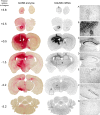
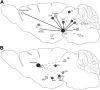
Neurogenetic disorders typically affect cells throughout the brain. Adeno-associated virus (AAV) vector-mediated transfer of a normal cDNA can correct the metabolic defects at the site of injection, but treatment of the entire brain requires widespread delivery of the normal gene and/or protein. Current methods require multiple injections for widespread distribution. However, some AAV vectors can be transported along neuronal pathways associated with the injected region. Thus, targeting widely dispersed systems in the CNS might be a pathway for gene dispersal from a limited number of sites. We tested this hypothesis in the ventral tegmental area (VTA), a region with numerous efferent and afferent projections. A single 1 mul injection resulted in transport of the vector genome to projection sites in distal parts of the brain. When compared with injections into the striatum, the VTA injection resulted in higher enzyme levels in more regions of the brain. The AAV-9 serotype vector was the most widely disseminated, but AAV-Rh.10 and AAV-1 were also transported after VTA injection. The effect on global lesions of a neurogenetic disease was tested in the mouse model of MPS VII (mucopolysaccharidosis VII), a lysosomal storage disorder. Widespread distribution of the vector genome after AAV-9 VTA injection resulted in even further distribution of the enzyme product, by secretion and uptake by surrounding cells, and complete correction of the storage lesions throughout the entire brain. This unprecedented level of correction from a single injection into the developed brain provides a potential strategy to correct a large volume of brain while minimizing the number of injections.
Figures


References
-
- Altman J. Autoradiographic and histological studies of postnatal neurogenesis. IV. Cell proliferation and migration in the anterior forebrain, with special reference to persisting neurogenesis in the olfactory bulb. J Comp Neurol. 1969;137:433–457. - PubMed
-
- Bartlett JS, Samulski RJ. Fluorescent viral vectors: a new technique for the pharmacological analysis of gene therapy. Nat Med. 1998;4:635–637. - PubMed
-
- Bartlett JS, Samulski RJ, McCown TJ. Selective and rapid uptake of adeno-associated virus type 2 in brain. Hum Gene Ther. 1998;9:1181–1186. - PubMed
-
- Beckstead RM, Domesick VB, Nauta WJ. Efferent connections of the substantia nigra and ventral tegmental area in the rat. Brain Res. 1979;175:191–217. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
