Characterization of heterotypic interaction effects in vitro to deconvolute global gene expression profiles in cancer
- PMID: 17868458
- PMCID: PMC2375029
- DOI: 10.1186/gb-2007-8-9-r191
Characterization of heterotypic interaction effects in vitro to deconvolute global gene expression profiles in cancer
Abstract
Background: Perturbations in cell-cell interactions are a key feature of cancer. However, little is known about the systematic effects of cell-cell interaction on global gene expression in cancer.
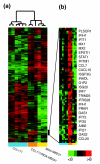
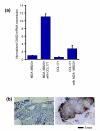
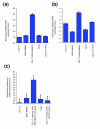
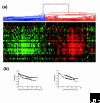
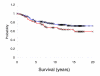
Results: We used an ex vivo model to simulate tumor-stroma interaction by systematically co-cultivating breast cancer cells with stromal fibroblasts and determined associated gene expression changes with cDNA microarrays. In the complex picture of epithelial-mesenchymal interaction effects, a prominent characteristic was an induction of interferon-response genes (IRGs) in a subset of cancer cells. In close proximity to these cancer cells, the fibroblasts secreted type I interferons, which, in turn, induced expression of the IRGs in the tumor cells. Paralleling this model, immunohistochemical analysis of human breast cancer tissues showed that STAT1, the key transcriptional activator of the IRGs, and itself an IRG, was expressed in a subset of the cancers, with a striking pattern of elevated expression in the cancer cells in close proximity to the stroma. In vivo, expression of the IRGs was remarkably coherent, providing a basis for segregation of 295 early-stage breast cancers into two groups. Tumors with high compared to low expression levels of IRGs were associated with significantly shorter overall survival; 59% versus 80% at 10 years (log-rank p = 0.001).
Conclusion: In an effort to deconvolute global gene expression profiles of breast cancer by systematic characterization of heterotypic interaction effects in vitro, we found that an interaction between some breast cancer cells and stromal fibroblasts can induce an interferon-response, and that this response may be associated with a greater propensity for tumor progression.
Figures


References
-
- Howlett AR, Bissell MJ. The influence of tissue microenvironment (stroma and extracellular matrix) on the development and function of mammary epithelium. Epithelial Cell Biol. 1993;2:79–89. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

