Comparison of protein coding gene contents of the fungal phyla Pezizomycotina and Saccharomycotina
- PMID: 17868481
- PMCID: PMC2045113
- DOI: 10.1186/1471-2164-8-325
Comparison of protein coding gene contents of the fungal phyla Pezizomycotina and Saccharomycotina
Abstract
Background: Several dozen fungi encompassing traditional model organisms, industrial production organisms and human and plant pathogens have been sequenced recently and their particular genomic features analysed in detail. In addition comparative genomics has been used to analyse specific sub groups of fungi. Notably, analysis of the phylum Saccharomycotina has revealed major events of evolution such as the recent genome duplication and subsequent gene loss. However, little has been done to gain a comprehensive comparative view to the fungal kingdom. We have carried out a computational genome wide comparison of protein coding gene content of Saccharomycotina and Pezizomycotina, which include industrially important yeasts and filamentous fungi, respectively.
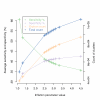
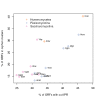
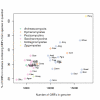
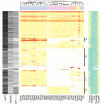
Results: Our analysis shows that based on genome redundancy, the traditional model organisms Saccharomyces cerevisiae and Neurospora crassa are exceptional among fungi. This can be explained by the recent genome duplication in S. cerevisiae and the repeat induced point mutation mechanism in N. crassa. Interestingly in Pezizomycotina a subset of protein families related to plant biomass degradation and secondary metabolism are the only ones showing signs of recent expansion. In addition, Pezizomycotina have a wealth of phylum specific poorly characterised genes with a wide variety of predicted functions. These genes are well conserved in Pezizomycotina, but show no signs of recent expansion. The genes found in all fungi except Saccharomycotina are slightly better characterised and predicted to encode mainly enzymes. The genes specific to Saccharomycotina are enriched in transcription and mitochondrion related functions. Especially mitochondrial ribosomal proteins seem to have diverged from those of Pezizomycotina. In addition, we highlight several individual gene families with interesting phylogenetic distributions.
Conclusion: Our analysis predicts that all Pezizomycotina unlike Saccharomycotina can potentially produce a wide variety of secondary metabolites and secreted enzymes and that the responsible gene families are likely to evolve fast. Both types of fungal products can be of commercial value, or on the other hand cause harm to humans. In addition, a great number of novel predicted and known enzymes are found from all fungi except Saccharomycotina. Therefore further studies and exploitation of fungal metabolism appears very promising.
Figures





References
-
- Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuveglise C, Talla E, Goffard N, Frangeul L, Aigle M, Anthouard V, Babour A, Barbe V, Barnay S, Blanchin S, Beckerich JM, Beyne E, Bleykasten C, Boisrame A, Boyer J, Cattolico L, Confanioleri F, De Daruvar A, Despons L, Fabre E, Fairhead C, Ferry-Dumazet H, Groppi A, Hantraye F, Hennequin C, Jauniaux N, Joyet P, Kachouri R, Kerrest A, Koszul R, Lemaire M, Lesur I, Ma L, Muller H, Nicaud JM, Nikolski M, Oztas S, Ozier-Kalogeropoulos O, Pellenz S, Potier S, Richard GF, Straub ML, Suleau A, Swennen D, Tekaia F, Wesolowski-Louvel M, Westhof E, Wirth B, Zeniou-Meyer M, Zivanovic I, Bolotin-Fukuhara M, Thierry A, Bouchier C, Caudron B, Scarpelli C, Gaillardin C, Weissenbach J, Wincker P, Souciet JL. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. - DOI - PubMed
-
- Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, Garcia JL, Garcia MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jimenez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafon A, Latge JP, Li W, Lord A, Lu C, Majoros WH, May GS, Miller BL, Mohamoud Y, Molina M, Monod M, Mouyna I, Mulligan S, Murphy L, O'Neil S, Paulsen I, Penalva MA, Pertea M, Price C, Pritchard BL, Quail MA, Rabbinowitsch E, Rawlins N, Rajandream MA, Reichard U, Renauld H, Robson GD, Rodriguez de Cordoba S, Rodriguez-Pena JM, Ronning CM, Rutter S, Salzberg SL, Sanchez M, Sanchez-Ferrero JC, Saunders D, Seeger K, Squares R, Squares S, Takeuchi M, Tekaia F, Turner G, Vazquez de Aldana CR, Weidman J, White O, Woodward J, Yu JH, Fraser C, Galagan JE, Asai K, Machida M, Hall N, Barrell B, Denning DW. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. - DOI - PubMed
-
- Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

