The GATA factor Serpent cross-regulates lozenge and u-shaped expression during Drosophila blood cell development
- PMID: 17869239
- PMCID: PMC2132443
- DOI: 10.1016/j.ydbio.2007.08.015
The GATA factor Serpent cross-regulates lozenge and u-shaped expression during Drosophila blood cell development
Abstract
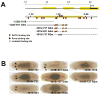
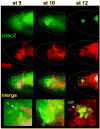
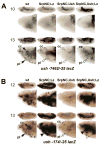
The Drosophila GATA factor Serpent interacts with the RUNX factor Lozenge to activate the crystal cell program, whereas SerpentNC binds the Friend of GATA protein U-shaped to limit crystal cell production. Here, we identified a lozenge minimal hematopoietic cis-regulatory module and showed that lozenge-lacZ reporter-gene expression was autoregulated by Serpent and Lozenge. We also showed that upregulation of u-shaped was delayed until after lozenge activation, consistent with our previous results that showed u-shaped expression in the crystal cell lineage is dependent on both Serpent and Lozenge. Together, these observations describe a feed forward regulatory motif, which controls the temporal expression of u-shaped. Finally, we showed that lozenge reporter-gene activity increased in a u-shaped mutant background and that forced expression of SerpentNC with U-shaped blocked lozenge- and u-shaped-lacZ reporter-gene activity. This is the first demonstration of GATA:FOG regulation of Runx and Fog gene expression. Moreover, these results identify components of a Serpent cross-regulatory sub-circuit that can modulate lozenge expression. Based on the sub-circuit design and the combinatorial control of crystal cell production, we present a model for the specification of a dynamic bi-potential regulatory state that contributes to the selection between a Lozenge-positive and Lozenge-negative state.
Figures




References
-
- Alfonso TB, Jones BW. gcm2 promotes glial cell differentiation and is required with glial cells missing for macrophage development in Drosophila. Dev Biol. 2002;248:369–383. - PubMed
-
- Anglin I, Passaniti A. Runx protein signaling in human cancers. Cancer Treat Res. 2004;119:189–215. - PubMed
-
- Bataille L, Auge B, Ferjoux G, Haenlin M, Waltzer L. Resolving embryonic blood cell fate choice in Drosophila: interplay of GCM and RUNX factors. Development. 2005;132:4635–4644. - PubMed
-
- Bernardoni R, Vivancos V, Giangrande A. glide/gcm is expressed and required in the scavenger cell lineage. Dev Biol. 1997;191:118–130. - PubMed
-
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

