Versatility in ligand recognition by LDL receptor family proteins: advances and frontiers
- PMID: 17870468
- PMCID: PMC2766800
- DOI: 10.1016/j.sbi.2007.08.017
Versatility in ligand recognition by LDL receptor family proteins: advances and frontiers
Abstract
Proteins of the low-density lipoprotein receptor family transport cholesterol-carrying particles into cells, clear protease-inhibitor complexes from the circulation, participate in biological signaling cascades, and even serve as viral receptors. These receptors utilize clusters of cysteine-rich LDL receptor type-A (LA) modules to bind many of their ligands. Recent structures show that these modules typically exhibit a characteristic binding mode to recognize their partners, relying primarily on electrostatic complementarity and avidity effects. The dominant contribution of electrostatic interactions with small interface areas in these complexes allows binding to be regulated by changes in pH via at least two distinct mechanisms. The structure of the subtilisin/kexin family protease PCSK9, a newly identified molecular partner of the LDLR also implicated in LDL-cholesterol homeostasis, also raises the possibility that the LDLR and its related family members may employ other strategies for pH-sensitive binding that have yet to be uncovered.
Figures
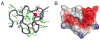



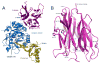
References
-
- Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. - PubMed
-
- Goldstein JL, Hobbs HH, Brown MS. Familial Hypercholesterolemia. In: Scriver CS, Beaudet AL, Sly WS, Vallee DV, editors. The Metabolic and Molecular Bases of Inherited Disease. 8. McGraw Hill Inc; 2001. pp. 2863–2913.
-
- Abifadel M, Varret M, Rabes JP, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. *This paper reports the identification of gain of function mutations in PCSK9 as a third cause of autosomal domainant hypercholesterolemia. - PubMed
-
- Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37:161–165. **This manuscript reports the important finding that loss-of-function mutations in PCSK9 are associated with hypocholesterolemia. Importantly, such individuals are apparently healthy, suggesting that pharmacologic inhibition of PCKS9 may be a viable new therapeutic approach for reducing LDL-cholesterol in humans. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

