Exoribonuclease R in Mycoplasma genitalium can carry out both RNA processing and degradative functions and is sensitive to RNA ribose methylation
- PMID: 17872508
- PMCID: PMC2040080
- DOI: 10.1261/rna.706207
Exoribonuclease R in Mycoplasma genitalium can carry out both RNA processing and degradative functions and is sensitive to RNA ribose methylation
Abstract
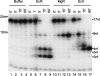
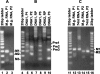
Mycoplasma genitalium, a small bacterium having minimal genome size, has only one identified exoribonuclease, RNase R (MgR). We have purified MgR to homogeneity, and compared its RNA degradative properties to those of its Escherichia coli homologs RNase R (EcR) and RNase II (EcII). MgR is active on a number of substrates including oligoribonucleotides, poly(A), rRNA, and precursors to tRNA. Unlike EcR, which degrades rRNA and pre-tRNA without formation of intermediate products, MgR appears sensitive to certain RNA structural features and forms specific products from these stable RNA substrates. The 3'-ends of two MgR degradation products of 23S rRNA were mapped by RT-PCR to positions 2499 and 2553, each being 1 nucleotide downstream of a 2'-O-methylation site. The sensitivity of MgR to ribose methylation is further demonstrated by the degradation patterns of 16S rRNA and a synthetic methylated oligoribonucleotide. Remarkably, MgR removes the 3'-trailer sequence from a pre-tRNA, generating product with the mature 3'-end more efficiently than EcII does. In contrast, EcR degrades this pre-tRNA without the formation of specific products. Our results suggest that MgR shares some properties of both EcR and EcII and can carry out a broad range of RNA processing and degradative functions.
Figures

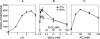

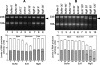


References
-
- Andrade, J.M., Cairrao, F., Arraiano, C.M. RNase R affects gene expression in stationary phase: Regulation of ompA. Mol. Microbiol. 2006;60:219–228. - PubMed
-
- Bollenbach, T.J., Lange, H., Gutierrez, R., Erhardt, M., Stern, D.B., Gagliardi, D. RNR1, a 3′–5′ exoribonuclease belonging to the RNR superfamily, catalyzes 3′ maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana . Nucleic Acids Res. 2005;33:2751–2763. doi: 10.1093/nar/gki576. - DOI - PMC - PubMed
-
- Cairrao, F., Cruz, A., Mori, H., Arraiano, C.M. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol. Microbiol. 2003;50:1349–1360. - PubMed
-
- Cannone, J.J., Subramanian, S., Schnare, M.N., Collett, J.R., D'Souza, L.M., Du, Y., Feng, B., Lin, N., Madabusi, L.V., Muller, K.M., et al. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002a;3:2. - PMC - PubMed
-
- Cannone, J.J., Subramanian, S., Schnare, M.N., Collett, J.R., D'Souza, L.M., Du, Y., Feng, B., Lin, N., Madabusi, L.V., Muller, K.M., et al. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs: Correction. BMC Bioinformatics. 2002b;3:15. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
