Role for the RecBCD recombination pathway for pilE gene variation in repair-proficient Neisseria gonorrhoeae
- PMID: 17873032
- PMCID: PMC2168704
- DOI: 10.1128/JB.00980-07
Role for the RecBCD recombination pathway for pilE gene variation in repair-proficient Neisseria gonorrhoeae
Abstract
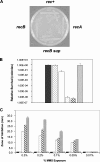
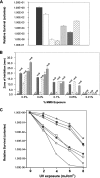
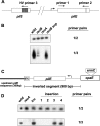
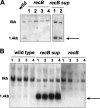
The role of the RecBCD recombination pathway in PilE antigenic variation in Neisseria gonorrhoeae is contentious and appears to be strain dependent. In this study, N. gonorrhoeae strain MS11 recB mutants were assessed for recombination/repair. MS11 recB mutants were found to be highly susceptible to DNA treatments that caused double-chain breaks and were severely impaired for growth; recB growth suppressor mutants arose at high frequencies. When the recombination/repair capacity of strain MS11 was compared to that of strains FA1090 and P9, innate differences were observed between the strains, with FA1090 and P9 rec(+) bacteria presenting pronounced recombination/repair defects. Consequently, MS11 recB mutants present a more robust phenotype than the other strains that were tested. In addition, MS11 recB mutants are also shown to be defective for pilE/pilS recombination. Moreover, pilE/pilS recombination is shown to proceed with gonococci that carry inverted pilE loci. Consequently, a novel RecBCD-mediated double-chain-break repair model for PilE antigenic variation is proposed.
Figures


Similar articles
-
Pilin antigenic variation occurs independently of the RecBCD pathway in Neisseria gonorrhoeae.J Bacteriol. 2009 Sep;191(18):5613-21. doi: 10.1128/JB.00535-09. Epub 2009 Jul 10. J Bacteriol. 2009. PMID: 19592592 Free PMC article.
-
RegF, an SspA homologue, regulates the expression of the Neisseria gonorrhoeae pilE gene.Res Microbiol. 1997 May;148(4):289-303. doi: 10.1016/S0923-2508(97)81585-9. Res Microbiol. 1997. PMID: 9765808
-
Antigenic variation of gonococcal pilin expression in vivo: analysis of the strain FA1090 pilin repertoire and identification of the pilS gene copies recombining with pilE during experimental human infection.Microbiology (Reading). 2001 Apr;147(Pt 4):839-849. doi: 10.1099/00221287-147-4-839. Microbiology (Reading). 2001. PMID: 11283280
-
Pilin gene variation in Neisseria gonorrhoeae: reassessing the old paradigms.FEMS Microbiol Rev. 2009 May;33(3):521-30. doi: 10.1111/j.1574-6976.2009.00171.x. FEMS Microbiol Rev. 2009. PMID: 19396954 Free PMC article. Review.
-
Neisseria gonorrhoeae: DNA Repair Systems and Their Role in Pathogenesis.Biochemistry (Mosc). 2022 Sep;87(9):965-982. doi: 10.1134/S0006297922090097. Biochemistry (Mosc). 2022. PMID: 36180987 Review.
Cited by
-
Mobile DNA in the pathogenic Neisseria.Microbiol Spectr. 2015 Feb;3(3):MDNA3-0015-2014. doi: 10.1128/microbiolspec.MDNA3-0015-2014. Microbiol Spectr. 2015. PMID: 25866700 Free PMC article.
-
Pilin antigenic variation occurs independently of the RecBCD pathway in Neisseria gonorrhoeae.J Bacteriol. 2009 Sep;191(18):5613-21. doi: 10.1128/JB.00535-09. Epub 2009 Jul 10. J Bacteriol. 2009. PMID: 19592592 Free PMC article.
-
Organization of the electron transfer chain to oxygen in the obligate human pathogen Neisseria gonorrhoeae: roles for cytochromes c4 and c5, but not cytochrome c2, in oxygen reduction.J Bacteriol. 2010 May;192(9):2395-406. doi: 10.1128/JB.00002-10. Epub 2010 Feb 12. J Bacteriol. 2010. PMID: 20154126 Free PMC article.
-
H-NS suppresses pilE intragenic transcription and antigenic variation in Neisseria gonorrhoeae.Microbiology (Reading). 2016 Jan;162(1):177-190. doi: 10.1099/mic.0.000199. Epub 2015 Oct 16. Microbiology (Reading). 2016. PMID: 26475082 Free PMC article.
-
Microbial antigenic variation mediated by homologous DNA recombination.FEMS Microbiol Rev. 2012 Sep;36(5):917-48. doi: 10.1111/j.1574-6976.2011.00321.x. Epub 2012 Jan 17. FEMS Microbiol Rev. 2012. PMID: 22212019 Free PMC article. Review.
References
-
- Bhat, K., C. P. Gibbs, O. Barrera, S. G. Morrison, F. Jahnig, A. Stern, E. M. Kupsch, T. F. Meyer, and J. Swanson. 1991. The opacity proteins of Neisseria gonorrhoeae strain MS11 are encoded by a family of 11 complete genes. Mol. Microbiol. 5:1889-1901. - PubMed
-
- Chaussee, M. S., J. Wilson, and S. A. Hill. 1999. Characterization of the recD gene of Neisseria gonorrhoeae MS11 and the effect of recD inactivation on pilin variation and DNA transformation. Microbiology 145:389-400. - PubMed
-
- Davidsen, T., E. K. Amundsen, E. A. Rodland, and T. Tonjum. 2007. DNA repair profiles of disease-associated isolates of Neisseria meningitidis. FEMS Immunol. Med. Microbiol. 49:243-251. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

