Crystallographic trapping in the rebeccamycin biosynthetic enzyme RebC
- PMID: 17873060
- PMCID: PMC2000502
- DOI: 10.1073/pnas.0707190104
Crystallographic trapping in the rebeccamycin biosynthetic enzyme RebC
Abstract
The biosynthesis of rebeccamycin, an antitumor compound, involves the remarkable eight-electron oxidation of chlorinated chromopyrrolic acid. Although one rebeccamycin biosynthetic enzyme is capable of generating low levels of the eight-electron oxidation product on its own, a second protein, RebC, is required to accelerate product formation and eliminate side reactions. However, the mode of action of RebC was largely unknown. Using crystallography, we have determined a likely function for RebC as a flavin hydroxylase, captured two snapshots of its dynamic catalytic cycle, and trapped a reactive molecule, a putative substrate, in its binding pocket. These studies strongly suggest that the role of RebC is to sequester a reactive intermediate produced by its partner protein and to react with it enzymatically, preventing its conversion to a suite of degradation products that includes, at low levels, the desired product.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


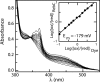
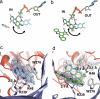
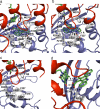
Comment in
-
Crystallography gets the jump on the enzymologists.Proc Natl Acad Sci U S A. 2007 Oct 2;104(40):15587-8. doi: 10.1073/pnas.0707843104. Epub 2007 Sep 26. Proc Natl Acad Sci U S A. 2007. PMID: 17898164 Free PMC article. No abstract available.
References
-
- Bush JA, Long BH, Catino JJ, Bradner WT, Tomita K. J Antibiot. 1987;40:668–678. - PubMed
-
- Staker BL, Feese MD, Cushman M, Pommier Y, Zembower D, Stewart L, Burgin AB. J Med Chem. 2005;48:2336–2345. - PubMed
-
- Prudhomme M. Eur J Med Chem. 2003;38:123–140. - PubMed
-
- Sanchez C, Butovich IA, Brana AF, Rohr J, Mendez C, Salas JA. Chem Biol. 2002;9:519–531. - PubMed
-
- Onaka H, Taniguchi S, Igarashi Y, Furumai T. Biosci Biotech Biochem. 2003;67:127–138. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

