Identification of a peripheral blood transcriptional biomarker panel associated with operational renal allograft tolerance
- PMID: 17873064
- PMCID: PMC2000539
- DOI: 10.1073/pnas.0705834104
Identification of a peripheral blood transcriptional biomarker panel associated with operational renal allograft tolerance
Abstract
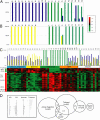
Long-term allograft survival generally requires lifelong immunosuppression (IS). Rarely, recipients display spontaneous "operational tolerance" with stable graft function in the absence of IS. The lack of biological markers of this phenomenon precludes identification of potentially tolerant patients in which IS could be tapered and hinders the development of new tolerance-inducing strategies. The objective of this study was to identify minimally invasive blood biomarkers for operational tolerance and use these biomarkers to determine the frequency of this state in immunosuppressed patients with stable graft function. Blood gene expression profiles from 75 renal-transplant patient cohorts (operational tolerance/acute and chronic rejection/stable graft function on IS) and 16 healthy individuals were analyzed. A subset of samples was used for microarray analysis where three-class comparison of the different groups of patients identified a "tolerant footprint" of 49 genes. These biomarkers were applied for prediction of operational tolerance by microarray and real-time PCR in independent test groups. Thirty-three of 49 genes correctly segregated tolerance and chronic rejection phenotypes with 99% and 86% specificity. The signature is shared with 1 of 12 and 5 of 10 stable patients on triple IS and low-dose steroid monotherapy, respectively. The gene signature suggests a pattern of reduced costimulatory signaling, immune quiescence, apoptosis, and memory T cell responses. This study identifies in the blood of kidney recipients a set of genes associated with operational tolerance that may have utility as a minimally invasive monitoring tool for guiding IS titration. Further validation of this tool for safe IS minimization in prospective clinical trials is warranted.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Opelz G. Transplantation. 1995;60:1220–1224. - PubMed
-
- Nankivell BJ, Borrows RJ, Fung CL, O'Connell PJ, Allen RD, Chapman JR. N Engl J Med. 2003;349:2326–2333. - PubMed
-
- Dantal J, Hourmant M, Cantarovich D, Giral M, Blancho G, Dreno B, JP S. Lancet. 1998;351:623–628. - PubMed
-
- Iman A, Rao M, Juneja R, Jacob CK. Natl Med J India. 2001;14:75–80. - PubMed
-
- Strober S. J Allergy Clin Immunol. 2000;106:S113–S114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

