INCURVATA2 encodes the catalytic subunit of DNA Polymerase alpha and interacts with genes involved in chromatin-mediated cellular memory in Arabidopsis thaliana
- PMID: 17873092
- PMCID: PMC2048701
- DOI: 10.1105/tpc.107.054130
INCURVATA2 encodes the catalytic subunit of DNA Polymerase alpha and interacts with genes involved in chromatin-mediated cellular memory in Arabidopsis thaliana
Abstract
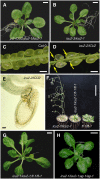
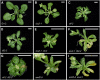
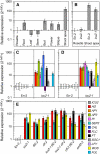
Cell type-specific gene expression patterns are maintained by the stable inheritance of transcriptional states through mitosis, requiring the action of multiprotein complexes that remodel chromatin structure. Genetic and molecular interactions between chromatin remodeling factors and components of the DNA replication machinery have been identified in Schizosaccharomyces pombe, indicating that some epigenetic marks are replicated simultaneously to DNA with the participation of the DNA replication complexes. This model of epigenetic inheritance might be extended to the plant kingdom, as we report here with the positional cloning and characterization of INCURVATA2 (ICU2), which encodes the putative catalytic subunit of the DNA polymerase alpha of Arabidopsis thaliana. The strong icu2-2 and icu2-3 insertional alleles caused fully penetrant zygotic lethality when homozygous and incompletely penetrant gametophytic lethality, probably because of loss of DNA polymerase activity. The weak icu2-1 allele carried a point mutation and caused early flowering, leaf incurvature, and homeotic transformations of sepals into carpels and of petals into stamens. Further genetic analyses indicated that ICU2 interacts with TERMINAL FLOWER2, the ortholog of HETEROCHROMATIN PROTEIN1 of animals and yeasts, and with the Polycomb group (PcG) gene CURLY LEAF. Another PcG gene, EMBRYONIC FLOWER2, was found to be epistatic to ICU2. Quantitative RT-PCR analyses indicated that a number of regulatory genes were derepressed in the icu2-1 mutant, including genes associated with flowering time, floral meristem, and floral organ identity.
Figures






References
-
- Ahmad, K., and Henikoff, S. (2002). Epigenetic consequences of nucleosome dynamics. Cell 111 281–284. - PubMed
-
- Ahmed, S., Saini, S., Arora, S., and Singh, J. (2001). Chromodomain protein Swi6-mediated role of DNA polymerase alpha in establishment of silencing in fission yeast. J. Biol. Chem. 276 47814–47821. - PubMed
-
- Ayyanathan, K., Lechner, M.S., Bell, P., Maul, G.G., Schultz, D.C., Yamada, Y., Tanaka, K., Torigoe, K., and Rauscher III, F.J. (2003). Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: A mammalian cell culture model of gene variegation. Genes Dev. 17 1855–1869. - PMC - PubMed
-
- Bannister, A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C., and Kouzarides, T. (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410 120–124. - PubMed
-
- Bastow, R., Mylne, J.S., Lister, C., Lippman, Z., Martienssen, R.A., and Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427 164–167. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

