Population genomics of human gene expression
- PMID: 17873874
- PMCID: PMC2683249
- DOI: 10.1038/ng2142
Population genomics of human gene expression
Abstract
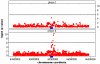
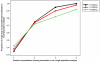
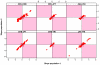
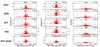
Genetic variation influences gene expression, and this variation in gene expression can be efficiently mapped to specific genomic regions and variants. Here we have used gene expression profiling of Epstein-Barr virus-transformed lymphoblastoid cell lines of all 270 individuals genotyped in the HapMap Consortium to elucidate the detailed features of genetic variation underlying gene expression variation. We find that gene expression is heritable and that differentiation between populations is in agreement with earlier small-scale studies. A detailed association analysis of over 2.2 million common SNPs per population (5% frequency in HapMap) with gene expression identified at least 1,348 genes with association signals in cis and at least 180 in trans. Replication in at least one independent population was achieved for 37% of cis signals and 15% of trans signals, respectively. Our results strongly support an abundance of cis-regulatory variation in the human genome. Detection of trans effects is limited but suggests that regulatory variation may be the key primary effect contributing to phenotypic variation in humans. We also explore several methodologies that improve the current state of analysis of gene expression variation.
Figures


Comment in
-
Pinpointing expression differences.Nat Genet. 2007 Oct;39(10):1175. doi: 10.1038/ng1007-1175. Nat Genet. 2007. PMID: 17898770 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

