Epidermal growth factor receptor (EGFR) is transcriptionally induced by the Y-box binding protein-1 (YB-1) and can be inhibited with Iressa in basal-like breast cancer, providing a potential target for therapy
- PMID: 17875215
- PMCID: PMC2242657
- DOI: 10.1186/bcr1767
Epidermal growth factor receptor (EGFR) is transcriptionally induced by the Y-box binding protein-1 (YB-1) and can be inhibited with Iressa in basal-like breast cancer, providing a potential target for therapy
Abstract
Introduction: Basal-like breast cancers (BLBCs) are very aggressive, and present serious clinical challenges as there are currently no targeted therapies available. We determined the regulatory role of Y-box binding protein-1 (YB-1) on epidermal growth factor receptor (EGFR) overexpression in BLBC, and the therapeutic potential of inhibiting EGFR. We pursued this in light of our recent work showing that YB-1 induces the expression of EGFR, a new BLBC marker.
Methods: Primary tumour tissues were evaluated for YB1 protein expression by immunostaining tissue microarrays, while copy number changes were assessed by comparative genomic hybridization (CGH). The ability of YB-1 to regulate EGFR was evaluated using luciferase reporter, chromatin immunoprecipitation (ChIP) and gel shift assays. The impact of Iressa on monolayer cell growth was measured using an ArrayScan VTI high-throughput analyser to count cell number, and colony formation in soft agar was used to measure anchorage-independent growth.
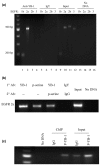
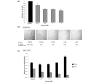
Results: YB-1 (27/37 or 73% of cases, P = 3.899 x 10(-4)) and EGFR (20/37 or 57.1% of cases, P = 9.206 x 10(-12)) are expressed in most cases of BLBC. However, they are not typically amplified in primary BLBC, suggesting overexpression owing to transcriptional activation. In support of this, we demonstrate that YB-1 promotes EGFR reporter activity. YB-1 specifically binds the EGFR promoter at two different YB-1-responsive elements (YREs) located at -940 and -968 using ChIP and gel shift assays in a manner that is dependent on the phosphorylation of S102 on YB-1. Inhibiting EGFR with Iressa suppressed the growth of SUM149 cells by approximately 40% in monolayer, independent of mutations in the receptor. More importantly anchorage-independent growth of BLBC cell lines was inhibited with combinations of Iressa and YB-1 suppression.
Conclusion: We have identified for the first time a causal link for the expression of EGFR in BLBC through the induction by YB-1 where it binds specifically to two distinguished YREs. Finally, inhibition of EGFR in combination with suppression of YB-1 presents a potential opportunity for therapy in BLBC.
Figures




Similar articles
-
Disruption of the Y-box binding protein-1 results in suppression of the epidermal growth factor receptor and HER-2.Cancer Res. 2006 May 1;66(9):4872-9. doi: 10.1158/0008-5472.CAN-05-3561. Cancer Res. 2006. PMID: 16651443
-
Profiling YB-1 target genes uncovers a new mechanism for MET receptor regulation in normal and malignant human mammary cells.Oncogene. 2009 Mar 19;28(11):1421-31. doi: 10.1038/onc.2008.485. Epub 2009 Jan 19. Oncogene. 2009. PMID: 19151767
-
Y-box binding protein-1 serine 102 is a downstream target of p90 ribosomal S6 kinase in basal-like breast cancer cells.Breast Cancer Res. 2008;10(6):R99. doi: 10.1186/bcr2202. Epub 2008 Nov 27. Breast Cancer Res. 2008. PMID: 19036157 Free PMC article.
-
An update of the mechanisms of resistance to EGFR-tyrosine kinase inhibitors in breast cancer: Gefitinib (Iressa) -induced changes in the expression and nucleo-cytoplasmic trafficking of HER-ligands (Review).Int J Mol Med. 2007 Jul;20(1):3-10. Int J Mol Med. 2007. PMID: 17549382 Review.
-
Y-box binding protein 1--a prognostic marker and target in tumour therapy.Eur J Cell Biol. 2014 Jan-Feb;93(1-2):61-70. doi: 10.1016/j.ejcb.2013.11.007. Epub 2013 Dec 27. Eur J Cell Biol. 2014. PMID: 24461929 Review.
Cited by
-
Y-box binding protein-1 promotes tumorigenesis and progression via the epidermal growth factor receptor/AKT pathway in spinal chordoma.Cancer Sci. 2019 Jan;110(1):166-179. doi: 10.1111/cas.13875. Epub 2018 Dec 19. Cancer Sci. 2019. PMID: 30426615 Free PMC article.
-
Inhibition of Transcription Induces Phosphorylation of YB-1 at Ser102 and Its Accumulation in the Nucleus.Cells. 2019 Dec 31;9(1):104. doi: 10.3390/cells9010104. Cells. 2019. PMID: 31906126 Free PMC article.
-
Expression of Y-box-binding protein YB-1 allows stratification into long- and short-term survivors of head and neck cancer patients.Br J Cancer. 2011 Dec 6;105(12):1864-73. doi: 10.1038/bjc.2011.491. Epub 2011 Nov 17. Br J Cancer. 2011. PMID: 22095225 Free PMC article.
-
Lapatinib in combination with radiation diminishes tumor regrowth in HER2+ and basal-like/EGFR+ breast tumor xenografts.Int J Radiat Oncol Biol Phys. 2010 Jun 1;77(2):575-81. doi: 10.1016/j.ijrobp.2009.12.063. Int J Radiat Oncol Biol Phys. 2010. PMID: 20457354 Free PMC article.
-
Prognostic role of YB-1 expression in breast cancer: a meta-analysis.Int J Clin Exp Med. 2015 Feb 15;8(2):1780-91. eCollection 2015. Int J Clin Exp Med. 2015. PMID: 25932106 Free PMC article.
References
-
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnson H, Hastie T, Eisen M, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. - DOI - PMC - PubMed
-
- Wu J, Lee C, Yokom D, Jiang H, Cheang MCU, Yorida E, Turbin D, Berquin IM, Mertens PR, Iftner T, et al. Disruption of the Y-box binding protein-1 (YB-1) results in suppression of the epidermal growth factor receptor and Her-2. Cancer Res. 2006;66:4872–4879. doi: 10.1158/0008-5472.CAN-05-3561. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

