NLR proteins: integral members of innate immunity and mediators of inflammatory diseases
- PMID: 17875812
- PMCID: PMC3256237
- DOI: 10.1189/jlb.0607402
NLR proteins: integral members of innate immunity and mediators of inflammatory diseases
Abstract
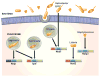
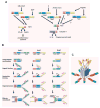
The innate immune system is the first line of defense against microorganisms and is conserved in plants and animals. The nucleotide-binding domain, leucine rich containing (NLR) protein family is a recent addition to the members of innate immunity effector molecules. These proteins are characterized by a central oligomerization domain, termed nucleotide-binding domain (NBD) and a protein interaction domain, leucine-rich repeats (LRRs) at the C terminus. It has been shown that NLR proteins are localized to the cytoplasm and recognize microbial products. To date, it is known that Nod1 and Nod2 detect bacterial cell wall components, whereas Ipaf and Naip detect bacterial flagellin, and NACHT/LRR/Pyrin 1 has been shown to detect anthrax lethal toxin. NLR proteins comprise a diverse protein family (over 20 in humans), indicating that NLRs have evolved to acquire specificity to various pathogenic microorganisms, thereby controlling host-pathogen interactions. Activation of NLR proteins results in inflammatory responses mediated by NF-kappaB, MAPK, or Caspase-1 activation, accompanied by subsequent secretion of proinflammatory cytokines. Mutations in several members of the NLR protein family have been linked to inflammatory diseases, suggesting these molecules play important roles in maintaining host-pathogen interactions and inflammatory responses. Therefore, understanding NLR signaling is important for the therapeutic intervention of various infectious and inflammatory diseases.
Figures



Similar articles
-
Genetic and Epigenetic Regulation of the Innate Immune Response to Gout.Immunol Invest. 2023 Apr;52(3):364-397. doi: 10.1080/08820139.2023.2168554. Epub 2023 Feb 6. Immunol Invest. 2023. PMID: 36745138 Review.
-
Heterotypic interactions among NACHT domains: implications for regulation of innate immune responses.Biochem J. 2004 Jul 1;381(Pt 1):213-9. doi: 10.1042/BJ20031506. Biochem J. 2004. PMID: 15107016 Free PMC article.
-
Roles of NOD1 (NLRC1) and NOD2 (NLRC2) in innate immunity and inflammatory diseases.Biosci Rep. 2012 Dec;32(6):597-608. doi: 10.1042/BSR20120055. Biosci Rep. 2012. PMID: 22908883 Free PMC article. Review.
-
NOD-LRR proteins: role in host-microbial interactions and inflammatory disease.Annu Rev Biochem. 2005;74:355-83. doi: 10.1146/annurev.biochem.74.082803.133347. Annu Rev Biochem. 2005. PMID: 15952891 Review.
-
Nods, Nalps and Naip: intracellular regulators of bacterial-induced inflammation.Cell Microbiol. 2003 Sep;5(9):581-92. doi: 10.1046/j.1462-5822.2003.00304.x. Cell Microbiol. 2003. PMID: 12925128 Review.
Cited by
-
NLRP7 deubiquitination by USP10 promotes tumor progression and tumor-associated macrophage polarization in colorectal cancer.J Exp Clin Cancer Res. 2021 Apr 10;40(1):126. doi: 10.1186/s13046-021-01920-y. J Exp Clin Cancer Res. 2021. PMID: 33838681 Free PMC article.
-
Adenosine-Induced NLRP11 in B Lymphoblasts Suppresses Human CD4+ T Helper Cell Responses.J Immunol Res. 2020 Aug 3;2020:1421795. doi: 10.1155/2020/1421795. eCollection 2020. J Immunol Res. 2020. PMID: 32832566 Free PMC article.
-
A crucial role of Flagellin in the induction of airway mucus production by Pseudomonas aeruginosa.PLoS One. 2012;7(7):e39888. doi: 10.1371/journal.pone.0039888. Epub 2012 Jul 2. PLoS One. 2012. PMID: 22768318 Free PMC article.
-
Muramyl dipeptide-based analogs as potential anticancer compounds: Strategies to improve selectivity, biocompatibility, and efficiency.Front Oncol. 2022 Sep 27;12:970967. doi: 10.3389/fonc.2022.970967. eCollection 2022. Front Oncol. 2022. PMID: 36237313 Free PMC article. Review.
-
NOD2 Versus MEFV: Differential Diagnosis of Yao Syndrome and Familial Mediterranean Fever.Rheumatol Immunol Res. 2021 Dec 31;2(4):233-239. doi: 10.2478/rir-2021-0032. eCollection 2021 Dec. Rheumatol Immunol Res. 2021. PMID: 36467985 Free PMC article.
References
-
- Medzhitov R, Janeway CA., Jr Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997;91:295–8. - PubMed
-
- Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–76. - PubMed
-
- Inohara N, Nunez G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat Rev Immunol. 2003;3:371–82. - PubMed
-
- Kobayashi KS, Eynon EE, Flavell RA. Intracellular debugging. Nat Immunol. 2003;4:652–4. - PubMed
-
- Belkhadir Y, Subramaniam R, Dangl JL. Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr Opin Plant Biol. 2004;7:391–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

