Complex formation of yeast Rev1 with DNA polymerase eta
- PMID: 17875922
- PMCID: PMC2169172
- DOI: 10.1128/MCB.01478-07
Complex formation of yeast Rev1 with DNA polymerase eta
Abstract
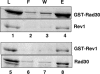
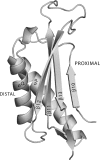
In Saccharomyces cerevisiae, Rev1 functions in translesion DNA synthesis (TLS) together with polymerase zeta (Pol zeta), comprised of the Rev3 catalytic and Rev7 accessory subunits. Rev1 plays an indispensable structural role in promoting Pol zeta function, and deletion of the Rev1-C terminal region that is involved in physical interactions with Rev3 inactivates Pol zeta function in TLS. In humans, however, Rev1 has been shown to physically interact with the Y-family polymerases Pol eta, Pol iota, and Pol kappa, and the Rev1 C terminus mediates these interactions. Since all the available genetic and biochemical evidence in yeast support the requirement of Rev1 as a structural element for Pol zeta and not for Pol eta, these observations have raised the possibility that in its structural role, Rev1 has diverged between yeast and humans. Here we show that although in yeast a stable Rev1-Pol eta complex can be formed, this complex formation involves the polymerase-associated domain of Rev1 and not the Rev1 C terminus as in humans. We also found that the DNA synthesis activity of Rev1 is enhanced in this complex. We discuss the implications of these and other observations for the possible divergence of Rev1's structural role between yeast and humans.
Figures


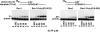

References
-
- Bailly, V., J. Lamb, P. Sung, S. Prakash, and L. Prakash. 1994. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 8:811-820. - PubMed
-
- Bailly, V., S. Lauder, S. Prakash, and L. Prakash. 1997. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 272:23360-23365. - PubMed
-
- Baynton, K., A. Bresson-Roy, and R. P. P. Fuchs. 1999. Distinct roles for Rev1p and Rev7p during translesion synthesis in Saccharomyces cerevisiae. Mol. Microbiol. 34:124-133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
