Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species
- PMID: 17877792
- PMCID: PMC2375034
- DOI: 10.1186/gb-2007-8-9-r196
Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species
Abstract
Background: The evolutionary history of organisms is expressed in phylogenetic trees. The most widely used phylogenetic trees describing the evolution of all organisms have been constructed based on single-gene phylogenies that, however, often produce conflicting results. Incongruence between phylogenetic trees can result from the violation of the orthology assumption and stochastic and systematic errors.
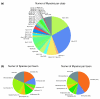
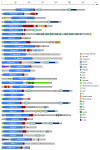
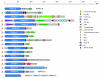
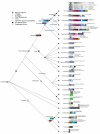
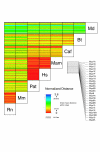
Results: Here, we have reconstructed the tree of eukaryotic life based on the analysis of 2,269 myosin motor domains from 328 organisms. All sequences were manually annotated and verified, and were grouped into 35 myosin classes, of which 16 have not been proposed previously. The resultant phylogenetic tree confirms some accepted relationships of major taxa and resolves disputed and preliminary classifications. We place the Viridiplantae after the separation of Euglenozoa, Alveolata, and Stramenopiles, we suggest a monophyletic origin of Entamoebidae, Acanthamoebidae, and Dictyosteliida, and provide evidence for the asynchronous evolution of the Mammalia and Fungi.
Conclusion: Our analysis of the myosins allowed combining phylogenetic information derived from class-specific trees with the information of myosin class evolution and distribution. This approach is expected to result in superior accuracy compared to single-gene or phylogenomic analyses because the orthology problem is resolved and a strong determinant not depending on any technical uncertainties is incorporated, the class distribution. Combining our analysis of the myosins with high quality analyses of other protein families, for example, that of the kinesins, could help in resolving still questionable dependencies at the origin of eukaryotic life.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

