Lightweight genome viewer: portable software for browsing genomics data in its chromosomal context
- PMID: 17877794
- PMCID: PMC2238324
- DOI: 10.1186/1471-2105-8-344
Lightweight genome viewer: portable software for browsing genomics data in its chromosomal context
Abstract
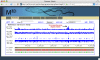
Background: Lightweight genome viewer (lwgv) is a web-based tool for visualization of sequence annotations in their chromosomal context. It performs most of the functions of larger genome browsers, while relying on standard flat-file formats and bypassing the database needs of most visualization tools. Visualization as an aide to discovery requires display of novel data in conjunction with static annotations in their chromosomal context. With database-based systems, displaying dynamic results requires temporary tables that need to be tracked for removal.
Results: lwgv simplifies the visualization of user-generated results on a local computer. The dynamic results of these analyses are written to transient files, which can import static content from a more permanent file. lwgv is currently used in many different applications, from whole genome browsers to single-gene RNAi design visualization, demonstrating its applicability in a large variety of contexts and scales.
Conclusion: lwgv provides a lightweight alternative to large genome browsers for visualizing biological annotations and dynamic analyses in their chromosomal context. It is particularly suited for applications ranging from short sequences to medium-sized genomes when the creation and maintenance of a large software and database infrastructure is not necessary or desired.
Figures


References
-
- Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, Hillman-Jackson J, Kuhn RM, Pedersen JS, Pohl A, Raney BJ, Rosenbloom KR, Siepel A, Smith KE, Sugnet CW, Sultan-Qurraie A, Thomas DJ, Trumbower H, Weber RJ, Weirauch M, Zweig AS, Haussler D, Kent WJ. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–8. doi: 10.1093/nar/gkj144. - DOI - PMC - PubMed
-
- Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Fitzgerald S, Fernandez-Banet J, Graf S, Haider S, Hammond M, Herrero J, Holland R, Howe K, Howe K, Johnson N, Kahari A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Melsopp C, Megy K, Meidl P, Ouverdin B, Parker A, Prlic A, Rice S, Rios D, Schuster M, Sealy I, Severin J, Slater G, Smedley D, Spudich G, Trevanion S, Vilella A, Vogel J, White S, Wood M, Cox T, Curwen V, Durbin R, Fernandez-Suarez XM, Flicek P, Kasprzyk A, Proctor G, Searle S, Smith J, Ureta-Vidal A, Birney E. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–7. doi: 10.1093/nar/gkl996. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

