Differential gene expression in an elite hybrid rice cultivar (Oryza sativa, L) and its parental lines based on SAGE data
- PMID: 17877838
- PMCID: PMC2077334
- DOI: 10.1186/1471-2229-7-49
Differential gene expression in an elite hybrid rice cultivar (Oryza sativa, L) and its parental lines based on SAGE data
Abstract
Background: It was proposed that differentially-expressed genes, aside from genetic variations affecting protein processing and functioning, between hybrid and its parents provide essential candidates for studying heterosis or hybrid vigor. Based our serial analysis of gene expression (SAGE) data from an elite Chinese super-hybrid rice (LYP9) and its parental cultivars (93-11 and PA64s) in three major tissue types (leaves, roots and panicles) at different developmental stages, we analyzed the transcriptome and looked for candidate genes related to rice heterosis.
Results: By using an improved strategy of tag-to-gene mapping and two recently annotated genome assemblies (93-11 and PA64s), we identified 10,268 additional high-quality tags, reaching a grand total of 20,595 together with our previous result. We further detected 8.5% and 5.9% physically-mapped genes that are differentially-expressed among the triad (in at least one of the three stages) with P-values less than 0.05 and 0.01, respectively. These genes distributed in 12 major gene expression patterns; among them, 406 up-regulated and 469 down-regulated genes (P < 0.05) were observed. Functional annotations on the identified genes highlighted the conclusion that up-regulated genes (some of them are known enzymes) in hybrid are mostly related to enhancing carbon assimilation in leaves and roots. In addition, we detected a group of up-regulated genes related to male sterility and 442 down-regulated genes related to signal transduction and protein processing, which may be responsible for rice heterosis.
Conclusion: We improved tag-to-gene mapping strategy by combining information from transcript sequences and rice genome annotation, and obtained a more comprehensive view on genes that related to rice heterosis. The candidates for heterosis-related genes among different genotypes provided new avenue for exploring the molecular mechanism underlying heterosis.
Figures


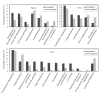
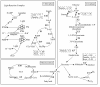
References
-
- Budak H. Understanding of Heterosis. KSU J Science and Engineering. 2002;5:69–75.
-
- Sun Q, Wu L, Ni Z, Meng F, Wang Z, Lin Z. Differential gene expression patterns in leaves between hybrids and their parental inbreds are correlated with heterosis in a wheat diallel cross. Plant Science (Oxford) 2004;166:651–657.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

