The E-protein Tcf4 interacts with Math1 to regulate differentiation of a specific subset of neuronal progenitors
- PMID: 17878293
- PMCID: PMC1978485
- DOI: 10.1073/pnas.0707456104
The E-protein Tcf4 interacts with Math1 to regulate differentiation of a specific subset of neuronal progenitors
Abstract
Proneural factors represent <10 transcriptional regulators required for specifying all of the different neurons of the mammalian nervous system. The mechanisms by which such a small number of factors creates this diversity are still unknown. We propose that proteins interacting with proneural factors confer such specificity. To test this hypothesis we isolated proteins that interact with Math1, a proneural transcription factor essential for the establishment of a neural progenitor population (rhombic lip) that gives rise to multiple hindbrain structures and identified the E-protein Tcf4. Interestingly, haploinsufficiency of TCF4 causes the Pitt-Hopkins mental retardation syndrome, underscoring the important role for this protein in neural development. To investigate the functional relevance of the Math1/Tcf4 interaction in vivo, we studied Tcf4(-/-) mice and found that they have disrupted pontine nucleus development. Surprisingly, this selective deficit occurs without affecting other rhombic lip-derived nuclei, despite expression of Math1 and Tcf4 throughout the rhombic lip. Importantly, deletion of any of the other E-protein-encoding genes does not have detectable effects on Math1-dependent neurons, suggesting a specialized role for Tcf4 in distinct neural progenitors. Our findings provide the first in vivo evidence for an exclusive function of dimers formed between a proneural basic helix-loop-helix factor and a specific E-protein, offering insight about the mechanisms underlying transcriptional programs that regulate development of the mammalian nervous system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
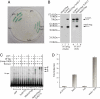



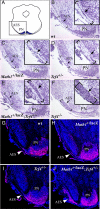
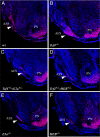
References
-
- Guillemot F. Curr Opin Cell Biol. 2005;17:639–647. - PubMed
-
- Ross SE, Greenberg ME, Stiles CD. Neuron. 2003;39:13–25. - PubMed
-
- Bertrand N, Castro DS, Guillemot F. Nat Rev Neurosci. 2002;3:517–530. - PubMed
-
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Cell. 1993;75:463–476. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

