Quantitative microarray profiling of DNA-binding molecules
- PMID: 17880081
- PMCID: PMC3066056
- DOI: 10.1021/ja0744899
Quantitative microarray profiling of DNA-binding molecules
Abstract
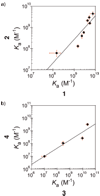
A high-throughput Cognate Site Identity (CSI) microarray platform interrogating all 524 800 10-base pair variable sites is correlated to quantitative DNase I footprinting data of DNA binding pyrrole-imidazole polyamides. An eight-ring hairpin polyamide programmed to target the 5 bp sequence 5'-TACGT-3' within the hypoxia response element (HRE) yielded a CSI microarray-derived sequence motif of 5'-WWACGT-3' (W = A,T). A linear beta-linked polyamide programmed to target a (GAA)3 repeat yielded a CSI microarray-derived sequence motif of 5'-AARAARWWG-3' (R = G,A). Quantitative DNase I footprinting of selected sequences from each microarray experiment enabled quantitative prediction of Ka values across the microarray intensity spectrum.
Figures








References
-
-
Im/Py targets G•C; Py/Py targets A•T and T•A; and Ct/Py targets T•A. Dervan PB, Edelson BS. Curr. Opin. Struct. Biol. 2003;13:284–299. Hsu CF, Phillips JW, Trauger JW, Farkas ME, Belitsky JM, Heckel A, Olenyuk BZ, Puckett JW, Wang CCC, Dervan PB. Tetrahedron. 2007;63:6146–6151.
-
-
- Trauger JW, Dervan PB. Methods Enzymol. 2001;340:450–466. - PubMed
-
-
(a) For a review on Protein Binding Microarrays (PBMs), see: Bulyk ML. Methods Enzymol. 2006;410:279–299. (b) For a review on fluorescence intercalator displacement (FID) assays, see: Tse WC, Boger DL. Acc. Chem. Res. 2004;37:61–69. (c) For the initial report of cognate site identifier (CSI) microarrays, see: Warren CL, Kratochvil NCS, Hauschild KE, Foister S, Brezinski ML, Dervan PB, Phillips GN, Ansari AZ. Proc. Natl. Acad. Sci. U.S.A. 2006;103:867–872.
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

