A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila
- PMID: 17880263
- PMCID: PMC1976628
- DOI: 10.1371/journal.pbio.0050251
A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila
Erratum in
- PLoS Biol. 2007 Nov;5(11):e291
Abstract
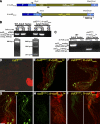
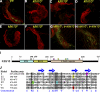
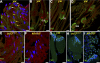
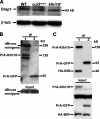
In both insects and mammals, spermatids eliminate their bulk cytoplasm as they undergo terminal differentiation. In Drosophila, this process of dramatic cellular remodeling requires apoptotic proteins, including caspases. To gain further insight into the regulation of caspases, we screened a large collection of sterile male flies for mutants that block effector caspase activation at the onset of spermatid individualization. Here, we describe the identification and characterization of a testis-specific, Cullin-3-dependent ubiquitin ligase complex that is required for caspase activation in spermatids. Mutations in either a testis-specific isoform of Cullin-3 (Cul3(Testis)), the small RING protein Roc1b, or a Drosophila orthologue of the mammalian BTB-Kelch protein Klhl10 all reduce or eliminate effector caspase activation in spermatids. Importantly, all three genes encode proteins that can physically interact to form a ubiquitin ligase complex. Roc1b binds to the catalytic core of Cullin-3, and Klhl10 binds specifically to a unique testis-specific N-terminal Cullin-3 (TeNC) domain of Cul3(Testis) that is required for activation of effector caspase in spermatids. Finally, the BIR domain region of the giant inhibitor of apoptosis-like protein dBruce is sufficient to bind to Klhl10, which is consistent with the idea that dBruce is a substrate for the Cullin-3-based E3-ligase complex. These findings reveal a novel role of Cullin-based ubiquitin ligases in caspase regulation.
Conflict of interest statement
Figures






Comment in
-
A protein complex restrains a homicidal enzyme during sperm differentiation.PLoS Biol. 2007 Oct;5(10):e279. doi: 10.1371/journal.pbio.0050279. Epub 2007 Sep 18. PLoS Biol. 2007. PMID: 20076648 Free PMC article. No abstract available.
References
-
- Abraham MC, Shaham S. Death without caspases, caspases without death. Trends Cell Biol. 2004;14:184–193. - PubMed
-
- Degterev A, Boyce M, Yuan J. A decade of caspases. Oncogene. 2003;22:8543–8567. - PubMed
-
- Lamkanfi M, Festjens N, Declercq W, Berghe TV, Vandenabeele P. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 2007;14:44–55. - PubMed
-
- Budihardjo I, Oliver H, Lutter M, Luo X, Wang X. Biochemical pathways of caspase activation during apoptosis. Annu Rev Cell Dev Biol. 1999;15:269–290. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

