Metabolic transcription analysis of engineered Escherichia coli strains that overproduce L-phenylalanine
- PMID: 17880710
- PMCID: PMC2089068
- DOI: 10.1186/1475-2859-6-30
Metabolic transcription analysis of engineered Escherichia coli strains that overproduce L-phenylalanine
Abstract
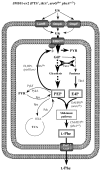
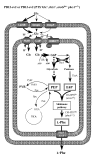
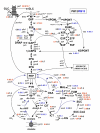
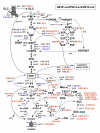
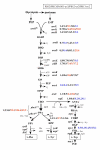
Background: The rational design of L-phenylalanine (L-Phe) overproducing microorganisms has been successfully achieved by combining different genetic strategies such as inactivation of the phosphoenolpyruvate: phosphotransferase transport system (PTS) and overexpression of key genes (DAHP synthase, transketolase and chorismate mutase-prephenate dehydratase), reaching yields of 0.33 (g-Phe/g-Glc), which correspond to 60% of theoretical maximum. Although genetic modifications introduced into the cell for the generation of overproducing organisms are specifically targeted to a particular pathway, these can trigger unexpected transcriptional responses of several genes. In the current work, metabolic transcription analysis (MTA) of both L-Phe overproducing and non-engineered strains using Real-Time PCR was performed, allowing the detection of transcriptional responses to PTS deletion and plasmid presence of genes related to central carbon metabolism. This MTA included 86 genes encoding enzymes of glycolysis, gluconeogenesis, pentoses phosphate, tricarboxylic acid cycle, fermentative and aromatic amino acid pathways. In addition, 30 genes encoding regulatory proteins and transporters for aromatic compounds and carbohydrates were also analyzed.
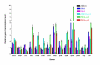
Results: MTA revealed that a set of genes encoding carbohydrate transporters (galP, mglB), gluconeogenic (ppsA, pckA) and fermentative enzymes (ldhA) were significantly induced, while some others were down-regulated such as ppc, pflB, pta and ackA, as a consequence of PTS inactivation. One of the most relevant findings was the coordinated up-regulation of several genes that are exclusively gluconeogenic (fbp, ppsA, pckA, maeB, sfcA, and glyoxylate shunt) in the best PTS- L-Phe overproducing strain (PB12-ev2). Furthermore, it was noticeable that most of the TCA genes showed a strong up-regulation in the presence of multicopy plasmids by an unknown mechanism. A group of genes exhibited transcriptional responses to both PTS inactivation and the presence of plasmids. For instance, acs-ackA, sucABCD, and sdhABCD operons were up-regulated in PB12 (PTS mutant that carries an arcB- mutation). The induction of these operons was further increased by the presence of plasmids in PB12-ev2. Some genes involved in the shikimate and specific aromatic amino acid pathways showed down-regulation in the L-Phe overproducing strains, might cause possible metabolic limitations in the shikimate pathway.
Conclusion: The identification of potential rate-limiting steps and the detection of transcriptional responses in overproducing microorganisms may suggest "reverse engineering" strategies for the further improvement of L-Phe production strains.
Figures
Similar articles
-
Metabolic engineering and protein directed evolution increase the yield of L-phenylalanine synthesized from glucose in Escherichia coli.Biotechnol Bioeng. 2004 Aug 20;87(4):516-24. doi: 10.1002/bit.20159. Biotechnol Bioeng. 2004. PMID: 15286989
-
[Co-expressions of phosphoenolpyruvate synthetase A (ppsA) and transketolase A (tktA) genes of Escherichia coli].Sheng Wu Gong Cheng Xue Bao. 2003 May;19(3):301-6. Sheng Wu Gong Cheng Xue Bao. 2003. PMID: 15969011 Chinese.
-
Acetate metabolism in Escherichia coli strains lacking phosphoenolpyruvate: carbohydrate phosphotransferase system; evidence of carbon recycling strategies and futile cycles.J Mol Microbiol Biotechnol. 2009;16(3-4):224-35. doi: 10.1159/000151219. Epub 2008 Aug 5. J Mol Microbiol Biotechnol. 2009. PMID: 18679018
-
Inactivation of the PTS as a Strategy to Engineer the Production of Aromatic Metabolites in Escherichia coli.J Mol Microbiol Biotechnol. 2015;25(2-3):195-208. doi: 10.1159/000380854. Epub 2015 Jul 9. J Mol Microbiol Biotechnol. 2015. PMID: 26159079 Review.
-
From scratch to value: engineering Escherichia coli wild type cells to the production of L-phenylalanine and other fine chemicals derived from chorismate.Appl Microbiol Biotechnol. 2007 Jun;75(4):739-49. doi: 10.1007/s00253-007-0931-y. Epub 2007 Apr 14. Appl Microbiol Biotechnol. 2007. PMID: 17435995 Review.
Cited by
-
Designing an Escherichia coli Strain for Phenylalanine Overproduction by Metabolic Engineering.Mol Biotechnol. 2017 May;59(4-5):168-178. doi: 10.1007/s12033-017-9999-5. Mol Biotechnol. 2017. PMID: 28374116
-
Metabolic engineering of microorganisms for production of aromatic compounds.Microb Cell Fact. 2019 Feb 26;18(1):41. doi: 10.1186/s12934-019-1090-4. Microb Cell Fact. 2019. PMID: 30808357 Free PMC article. Review.
-
Aggregatibacter actinomycetemcomitans arcB influences hydrophobic properties, biofilm formation and adhesion to hydroxyapatite.Braz J Microbiol. 2009 Jul;40(3):550-62. doi: 10.1590/S1517-838220090003000018. Epub 2009 Sep 1. Braz J Microbiol. 2009. PMID: 24031399 Free PMC article.
-
Protocell design through modular compartmentalization.J R Soc Interface. 2013 Aug 7;10(87):20130496. doi: 10.1098/rsif.2013.0496. Print 2013 Oct 6. J R Soc Interface. 2013. PMID: 23925982 Free PMC article.
-
Coutilization of glucose and glycerol enhances the production of aromatic compounds in an Escherichia coli strain lacking the phosphoenolpyruvate: carbohydrate phosphotransferase system.Microb Cell Fact. 2008 Jan 22;7:1. doi: 10.1186/1475-2859-7-1. Microb Cell Fact. 2008. PMID: 18211716 Free PMC article.
References
-
- Sprenger GA. From scratch to value: engineering Escherichia coli wild type cells to the production of L-phenylalanine and other fine chemicals derived from chorismate. Appl Microbiol Biotechnol. 2007 - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

