Structural biology of membrane-intrinsic beta-barrel enzymes: sentinels of the bacterial outer membrane
- PMID: 17880914
- PMCID: PMC5007122
- DOI: 10.1016/j.bbamem.2007.07.021
Structural biology of membrane-intrinsic beta-barrel enzymes: sentinels of the bacterial outer membrane
Abstract
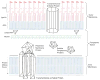
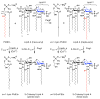
The outer membranes of Gram-negative bacteria are replete with integral membrane proteins that exhibit antiparallel beta-barrel structures, but very few of these proteins function as enzymes. In Escherichia coli, only three beta-barrel enzymes are known to exist in the outer membrane; these are the phospholipase OMPLA, the protease OmpT, and the phospholipidColon, two colonslipid A palmitoyltransferase PagP, all of which have been characterized at the structural level. Structural details have also emerged for the outer membrane beta-barrel enzyme PagL, a lipid A 3-O-deacylase from Pseudomonas aeruginosa. Lipid A can be further modified in the outer membrane by two beta-barrel enzymes of unknown structure; namely, the Salmonella enterica 3'-acyloxyacyl hydrolase LpxR, and the Rhizobium leguminosarum oxidase LpxQ, which employs O(2) to convert the proximal glucosamine unit of lipid A into 2-aminogluconate. Structural biology now indicates how beta-barrel enzymes can function as sentinels that remain dormant when the outer membrane permeability barrier is intact. Host immune defenses and antibiotics that perturb this barrier can directly trigger beta-barrel enzymes in the outer membrane. The ensuing adaptive responses occur instantaneously and rapidly outpace other signal transduction mechanisms that similarly function to restore the outer membrane permeability barrier.
Figures
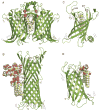
Similar articles
-
E. coli outer membrane and interactions with OmpLA.Biophys J. 2014 Jun 3;106(11):2493-502. doi: 10.1016/j.bpj.2014.04.024. Biophys J. 2014. PMID: 24896129 Free PMC article.
-
Solution structure and dynamics of the outer membrane enzyme PagP by NMR.Proc Natl Acad Sci U S A. 2002 Oct 15;99(21):13560-5. doi: 10.1073/pnas.212344499. Epub 2002 Sep 30. Proc Natl Acad Sci U S A. 2002. PMID: 12357033 Free PMC article.
-
Structure and function of bacterial outer membrane proteins: barrels in a nutshell.Mol Microbiol. 2000 Jul;37(2):239-53. doi: 10.1046/j.1365-2958.2000.01983.x. Mol Microbiol. 2000. PMID: 10931321 Review.
-
The lipid A palmitoyltransferase PagP: molecular mechanisms and role in bacterial pathogenesis.Mol Microbiol. 2005 Aug;57(4):900-12. doi: 10.1111/j.1365-2958.2005.04711.x. Mol Microbiol. 2005. PMID: 16091033 Review.
-
The versatile beta-barrel membrane protein.Curr Opin Struct Biol. 2003 Aug;13(4):404-11. doi: 10.1016/s0959-440x(03)00099-x. Curr Opin Struct Biol. 2003. PMID: 12948769 Review.
Cited by
-
Bacterial Sphingomyelinases and Phospholipases as Virulence Factors.Microbiol Mol Biol Rev. 2016 Jun 15;80(3):597-628. doi: 10.1128/MMBR.00082-15. Print 2016 Sep. Microbiol Mol Biol Rev. 2016. PMID: 27307578 Free PMC article. Review.
-
High-resolution structure prediction of β-barrel membrane proteins.Proc Natl Acad Sci U S A. 2018 Feb 13;115(7):1511-1516. doi: 10.1073/pnas.1716817115. Epub 2018 Jan 29. Proc Natl Acad Sci U S A. 2018. PMID: 29378944 Free PMC article.
-
A knowledge-based potential highlights unique features of membrane α-helical and β-barrel protein insertion and folding.Protein Sci. 2012 Jan;21(1):50-62. doi: 10.1002/pro.758. Epub 2011 Nov 23. Protein Sci. 2012. PMID: 22031179 Free PMC article.
-
Infection and distribution of Candidatus Liberibacter asiaticus in citrus plants and psyllid vectors at the cellular level.Microb Biotechnol. 2022 Apr;15(4):1221-1234. doi: 10.1111/1751-7915.13914. Epub 2021 Sep 1. Microb Biotechnol. 2022. PMID: 34469634 Free PMC article.
-
Mutant Alleles of lptD Increase the Permeability of Pseudomonas aeruginosa and Define Determinants of Intrinsic Resistance to Antibiotics.Antimicrob Agents Chemother. 2015 Nov 23;60(2):845-54. doi: 10.1128/AAC.01747-15. Print 2016 Feb. Antimicrob Agents Chemother. 2015. PMID: 26596941 Free PMC article.
References
-
- Snijder HJ, Ubarretxena-Belandia I, Blaauw M, Kalk KH, Verheij HM, Egmond MR, Dekker N, Dijkstra BW. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature. 1999;401:717–721. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

