Evidence that base stacking potential in annealed 3' overhangs determines polymerase utilization in yeast nonhomologous end joining
- PMID: 17881298
- PMCID: PMC2190084
- DOI: 10.1016/j.dnarep.2007.07.018
Evidence that base stacking potential in annealed 3' overhangs determines polymerase utilization in yeast nonhomologous end joining
Abstract
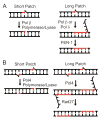
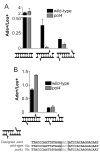
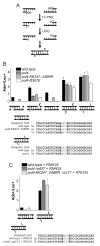
Nonhomologous end joining (NHEJ) directly rejoins DNA double-strand breaks (DSBs) when recombination is not possible. In Saccharomyces cerevisiae, the DNA polymerase Pol4 is required for gap filling when a short 3' overhang must prime DNA synthesis. Here, we examined further end variations to test specific hypotheses regarding Pol4 usage in NHEJ in vivo. Surprisingly, Pol4 dependence at 3' overhangs was reduced when a nonhomologous 5' flap nucleotide was present across from the gap, even though the mismatched nucleotide was corrected, not incorporated. In contrast, a gap with a 5' deoxyribosephosphate (dRP) was as Pol4-dependent as a gap with a 5' phosphate, demonstrating the importance of the downstream base in relaxing the Pol4 requirement. Combined with prior observations of Pol4-independent NHEJ of nicks with 5' hydroxyls, we suggest that base stacking interactions across the broken strands can stabilize a joint, allowing another polymerase to substitute for Pol4. This model predicts that a unique function of Pol4 is to actively stabilize template strands that lack stacking continuity. We also explored whether NHEJ end processing can occur via short- and long-patch pathways analogous to base excision repair. Results demonstrated that 5' dRPs could be removed in the absence of Pol4 lyase activity. The 5' flap endonuclease Rad27 was not required for repair in this or any situation tested, indicating that still other NHEJ 5' nucleases must exist.
Figures
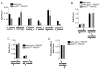
Similar articles
-
DNA joint dependence of pol X family polymerase action in nonhomologous end joining.J Biol Chem. 2005 Aug 12;280(32):29030-7. doi: 10.1074/jbc.M505277200. Epub 2005 Jun 17. J Biol Chem. 2005. PMID: 15964833
-
Pol3 is involved in nonhomologous end-joining in Saccharomyces cerevisiae.DNA Repair (Amst). 2008 Sep 1;7(9):1531-41. doi: 10.1016/j.dnarep.2008.05.008. Epub 2008 Jul 7. DNA Repair (Amst). 2008. PMID: 18606574
-
Role of the yeast DNA repair protein Nej1 in end processing during the repair of DNA double strand breaks by non-homologous end joining.DNA Repair (Amst). 2015 Jul;31:1-10. doi: 10.1016/j.dnarep.2015.04.003. Epub 2015 Apr 21. DNA Repair (Amst). 2015. PMID: 25942368 Free PMC article.
-
Tying up loose ends: nonhomologous end-joining in Saccharomyces cerevisiae.Mutat Res. 2000 Jun 30;451(1-2):71-89. doi: 10.1016/s0027-5107(00)00041-5. Mutat Res. 2000. PMID: 10915866 Review.
-
Consider the workhorse: Nonhomologous end-joining in budding yeast.Biochem Cell Biol. 2016 Oct;94(5):396-406. doi: 10.1139/bcb-2016-0001. Epub 2016 Mar 31. Biochem Cell Biol. 2016. PMID: 27240172 Free PMC article. Review.
Cited by
-
Nonhomologous DNA end-joining for repair of DNA double-strand breaks.J Biol Chem. 2018 Jul 6;293(27):10512-10523. doi: 10.1074/jbc.TM117.000374. Epub 2017 Dec 14. J Biol Chem. 2018. PMID: 29247009 Free PMC article. Review.
-
Repair of double-strand breaks by end joining.Cold Spring Harb Perspect Biol. 2013 May 1;5(5):a012757. doi: 10.1101/cshperspect.a012757. Cold Spring Harb Perspect Biol. 2013. PMID: 23637284 Free PMC article. Review.
-
Polymerases in nonhomologous end joining: building a bridge over broken chromosomes.Antioxid Redox Signal. 2011 Jun 15;14(12):2509-19. doi: 10.1089/ars.2010.3429. Epub 2010 Oct 28. Antioxid Redox Signal. 2011. PMID: 20649463 Free PMC article. Review.
-
The flexible and iterative steps within the NHEJ pathway.Prog Biophys Mol Biol. 2023 Jul-Aug;180-181:105-119. doi: 10.1016/j.pbiomolbio.2023.05.001. Epub 2023 May 5. Prog Biophys Mol Biol. 2023. PMID: 37150451 Free PMC article.
-
Structural basis for a novel mechanism of DNA bridging and alignment in eukaryotic DSB DNA repair.EMBO J. 2015 Apr 15;34(8):1126-42. doi: 10.15252/embj.201489643. Epub 2015 Mar 11. EMBO J. 2015. PMID: 25762590 Free PMC article.
References
-
- Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. - PubMed
-
- Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. - PubMed
-
- Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell. 2001;8:1105–1115. - PubMed
-
- Wilson TE, Grawunder U, Lieber MR. Yeast DNA ligase IV mediates nonhomologous DNA end joining. Nature. 1997;388:495–498. - PubMed
-
- Coquerelle T, Bopp A, Kessler B, Hagen U. Strand breaks and K’ end-groups in DNA of irradiated thymocytes. Int J Radiat Biol Relat Stud Phys Chem Med. 1973;24:397–404. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

