Human ribosomal protein S13 regulates expression of its own gene at the splicing step by a feedback mechanism
- PMID: 17881366
- PMCID: PMC2095825
- DOI: 10.1093/nar/gkm701
Human ribosomal protein S13 regulates expression of its own gene at the splicing step by a feedback mechanism
Abstract
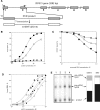
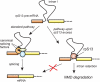
The expression of ribosomal protein (rp) genes is regulated at multiple levels. In yeast, two genes are autoregulated by feedback effects of the protein on pre-mRNA splicing. Here, we have investigated whether similar mechanisms occur in eukaryotes with more complicated and highly regulated splicing patterns. Comparisons of the sequences of ribosomal protein S13 gene (RPS13) among mammals and birds revealed that intron 1 is more conserved than the other introns. Transfection of HEK 293 cells with a minigene-expressing ribosomal protein S13 showed that the presence of intron 1 reduced expression by a factor of four. Ribosomal protein S13 was found to inhibit excision of intron 1 from rpS13 pre-mRNA fragment in vitro. This protein was shown to be able to specifically bind the fragment and to confer protection against ribonuclease cleavage at sequences near the 5' and 3' splice sites. The results suggest that overproduction of rpS13 in mammalian cells interferes with splicing of its own pre-mRNA by a feedback mechanism.
Figures

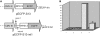


Similar articles
-
[Human ribosomal protein S13 inhibits splicing of the own pre-mRNA].Mol Biol (Mosk). 2007 Jan-Feb;41(1):51-8. Mol Biol (Mosk). 2007. PMID: 17380891 Russian.
-
[Human ribosomal protein S16 inhibites excision of the first intron from its own].Mol Biol (Mosk). 2010 Jan-Feb;44(1):90-7. Mol Biol (Mosk). 2010. PMID: 20198863 Russian.
-
[Ribosomal protein binding with the first intron of the human ribosomal protein S26 pre-mRNA stimulates its interaction with proteins extracted from Hela cells].Mol Biol (Mosk). 2002 May-Jun;36(3):503-10. Mol Biol (Mosk). 2002. PMID: 12068637 Russian.
-
Arabidopsis intron mutations and pre-mRNA splicing.Plant J. 1996 Nov;10(5):771-80. doi: 10.1046/j.1365-313x.1996.10050771.x. Plant J. 1996. PMID: 8953241 Review.
-
Pre-mRNA splicing: a complex picture in higher definition.Trends Biochem Sci. 2008 Jun;33(6):243-6. doi: 10.1016/j.tibs.2008.04.004. Epub 2008 May 9. Trends Biochem Sci. 2008. PMID: 18472266 Review.
Cited by
-
Specialized ribosomes: a new frontier in gene regulation and organismal biology.Nat Rev Mol Cell Biol. 2012 May 23;13(6):355-69. doi: 10.1038/nrm3359. Nat Rev Mol Cell Biol. 2012. PMID: 22617470 Free PMC article. Review.
-
Mutations in RPS19 may affect ribosome function and biogenesis in Diamond Blackfan anemia.FEBS Open Bio. 2022 Jul;12(7):1419-1434. doi: 10.1002/2211-5463.13444. Epub 2022 Jun 6. FEBS Open Bio. 2022. PMID: 35583751 Free PMC article.
-
Ribosomal proteins in hepatocellular carcinoma: mysterious but promising.Cell Biosci. 2024 Nov 1;14(1):133. doi: 10.1186/s13578-024-01316-3. Cell Biosci. 2024. PMID: 39487553 Free PMC article. Review.
-
Localization and Functional Roles of Components of the Translation Apparatus in the Eukaryotic Cell Nucleus.Cells. 2021 Nov 19;10(11):3239. doi: 10.3390/cells10113239. Cells. 2021. PMID: 34831461 Free PMC article. Review.
-
Auto-regulatory feedback by RNA-binding proteins.J Mol Cell Biol. 2019 Oct 25;11(10):930-939. doi: 10.1093/jmcb/mjz043. J Mol Cell Biol. 2019. PMID: 31152582 Free PMC article. Review.
References
-
- Ferreira-Cerca S, Poll G, Gleizes PE, Tschochner H, Milkereit P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell. 2005;20:263–275. - PubMed
-
- Bulygin K, Chavatte L, Frolova L, Karpova G, Favre A. The first position of a codon placed in the A site of the human 80S ribosome contacts nucleotide C1696 of the 18S rRNA as well as proteins S2, S3, S3a, S30, and S15. Biochemistry. 2005;44:2153–2162. - PubMed
-
- Wool IG. Extraribosomal functions of ribosomal proteins. Trends Biochem. Sci. 1996;21:164–165. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

