The role of peptide motifs in the evolution of a protein network
- PMID: 17881369
- PMCID: PMC2095796
- DOI: 10.1093/nar/gkm692
The role of peptide motifs in the evolution of a protein network
Abstract
Naturally occurring proteins in cellular networks often share peptide motifs. These motifs have been known to play a pivotal role in protein interactions among the components of a network. However, it remains unknown how these motifs have contributed to the evolution of the protein network. Here we addressed this issue by a synthetic biology approach. Through the motif programming method, we have constructed an artificial protein library by mixing four peptide motifs shared among the Bcl-2 family proteins that positively or negatively regulate the apoptosis networks. We found one strong pro-apoptotic protein, d29, and two proteins having moderate, but unambiguous anti-apoptotic functions, a10 and d16, from the 28 tested clones. Thus both the pro- and anti-apoptotic modulators were present in the library, demonstrating that functional proteins with opposing effects can emerge from a single pool prepared from common motifs. Motif programming studies have exhibited that the annotated function of the motifs were significantly influenced by the context that the motifs embedded. The results further revealed that reshuffling of a set of motifs realized the promiscuous state of protein, from which disparate functions could emerge. Our finding suggests that motifs contributed to the plastic evolvability of the protein network.
Figures
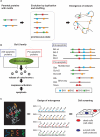


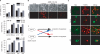
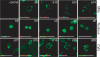
Similar articles
-
Motif programming: a microgene-based method for creating synthetic proteins containing multiple functional motifs.Nucleic Acids Res. 2007;35(6):e38. doi: 10.1093/nar/gkm017. Epub 2007 Feb 7. Nucleic Acids Res. 2007. PMID: 17287291 Free PMC article.
-
Synthesis of functional proteins by mixing peptide motifs.Chem Biol. 2004 Jun;11(6):765-73. doi: 10.1016/j.chembiol.2004.03.032. Chem Biol. 2004. PMID: 15217610
-
Sequence analysis shows that Lifeguard belongs to a new evolutionarily conserved cytoprotective family.Int J Mol Med. 2006 Oct;18(4):729-34. Int J Mol Med. 2006. PMID: 16964429
-
Natural and artificial peptide motifs: their origins and the application of motif-programming.Chem Soc Rev. 2010 Jan;39(1):117-26. doi: 10.1039/b719081f. Epub 2009 Sep 2. Chem Soc Rev. 2010. PMID: 20023842 Review.
-
The mechanism of Ca2+-dependent recognition of Alix by ALG-2: insights from X-ray crystal structures.Biochem Soc Trans. 2009 Feb;37(Pt 1):190-4. doi: 10.1042/BST0370190. Biochem Soc Trans. 2009. PMID: 19143629 Review.
Cited by
-
Unique N-glycosylation signatures in Aβ oligomer-and lipopolysaccharide-activated human iPSC-derived microglia.Res Sq [Preprint]. 2024 Nov 19:rs.3.rs-5308977. doi: 10.21203/rs.3.rs-5308977/v1. Res Sq. 2024. Update in: Sci Rep. 2025 Apr 10;15(1):12348. doi: 10.1038/s41598-025-96596-1. PMID: 39606433 Free PMC article. Updated. Preprint.
-
Antimicrobial peptide developed with machine learning sequence optimization targets drug resistant Staphylococcus aureus in mice.J Clin Invest. 2025 Apr 22;135(12):e185430. doi: 10.1172/JCI185430. eCollection 2025 Jun 16. J Clin Invest. 2025. PMID: 40261713 Free PMC article.
-
Phylogenetic identification of bacterial MazF toxin protein motifs among probiotic strains and foodborne pathogens and potential implications of engineered probiotic intervention in food.Cell Biosci. 2012 Nov 27;2(1):39. doi: 10.1186/2045-3701-2-39. Cell Biosci. 2012. PMID: 23186337 Free PMC article.
-
Unique N-glycosylation signatures in human iPSC derived microglia activated by Aβ oligomer and lipopolysaccharide.Sci Rep. 2025 Apr 10;15(1):12348. doi: 10.1038/s41598-025-96596-1. Sci Rep. 2025. PMID: 40210651 Free PMC article.
-
In Silico-Based Design of a Hybrid Peptide with Antimicrobial Activity against Multidrug-Resistant Pseudomonas aeruginosa Using a Spider Toxin Peptide.Toxins (Basel). 2023 Nov 23;15(12):668. doi: 10.3390/toxins15120668. Toxins (Basel). 2023. PMID: 38133172 Free PMC article.
References
-
- Kitano H. Biological robustness. Nat. Rev. Genet. 2004;5:826–837. - PubMed
-
- Stelling J, Sauer U, Szallasi Z, Doyle FJ, III, Doyle J. Robustness of cellular functions. Cell. 2004;118:675–685. - PubMed
-
- Patthy L. Evolution of the proteases of blood coagulation and fibrinolysis by assembly from modules. Cell. 1985;41:657–663. - PubMed
-
- King N, Hittinger CT, Carroll SB. Evolution of key cell signaling and adhesion protein families predates animal origins. Science. 2003;301:361–363. - PubMed

