Divergence and recombination of clinical herpes simplex virus type 2 isolates
- PMID: 17881457
- PMCID: PMC2169075
- DOI: 10.1128/JVI.01310-07
Divergence and recombination of clinical herpes simplex virus type 2 isolates
Abstract
Herpes simplex virus type 2 (HSV-2) infects the genital mucosa and is one of the most common sexually transmitted viruses. Here we sequenced a segment comprising 3.5% of the HSV-2 genome, including genes coding for glycoproteins G, I, and E, from 27 clinical isolates from Tanzania, 10 isolates from Norway, and 10 isolates from Sweden. The sequence variation was low compared to that described for clinical HSV-1 isolates, with an overall similarity of 99.6% between the two most distant HSV-2 isolates. Phylogenetic analysis revealed a divergence into at least two genogroups arbitrarily designated A and B, supported by high bootstrap values and evolutionarily separated at the root. Genogroup A contained isolates collected in Tanzania, and genogroup B contained isolates collected in Tanzania and Scandinavia, implying that the genetic variability of HSV-2 is higher in Tanzania than in Scandinavia. Recombination network analysis and bootscan analysis revealed a complex pattern of phylogenetically conflicting informative sites in the sequence alignments. These signals were present in synonymous and nonsynonymous sites in all three genes and were not accumulated in specific regions, observations arguing against positive selection. Since the PHI test applied solely to synonymous sites revealed a high statistical probability of recombination, we suggest as a novel finding that homologous recombination is, as reported earlier for HSV-1 and varicella-zoster virus, a prominent feature in the evolution of HSV-2.
Figures

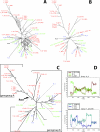
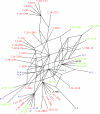
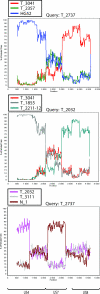
References
-
- Aurelius, E., B. Johansson, B. Skoldenberg, and M. Forsgren. 1993. Encephalitis in immunocompetent patients due to herpes simplex virus type 1 or 2 as determined by type-specific polymerase chain reaction and antibody assays of cerebrospinal fluid. J. Med. Virol. 39:179-186. - PubMed
-
- Bowden, R., H. Sakaoka, P. Donnelly, and R. Ward. 2004. High recombination rate in herpes simplex virus type 1 natural populations suggests significant co-infection. Infect. Genet. Evol. 4:115-123. - PubMed
-
- Brown, S. M., and D. A. Ritchie. 1975. Genetic studies with herpes simplex virus type 1. Analysis of mixed plaque-forming virus and its bearing on genetic recombination. Virology 64:32-42. - PubMed
-
- Brown, S. M., J. H. Subak-Sharpe, J. Harland, and A. R. MacLean. 1992. Analysis of intrastrain recombination in herpes simplex virus type 1 strain 17 and herpes simplex virus type 2 strain HG52 using restriction endonuclease sites as unselected markers and temperature-sensitive lesions as selected markers. J. Gen. Virol. 73:293-301. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

