Specificity of RGS10A as a key component in the RANKL signaling mechanism for osteoclast differentiation
- PMID: 17881498
- PMCID: PMC3587975
- DOI: 10.1242/jcs.008300
Specificity of RGS10A as a key component in the RANKL signaling mechanism for osteoclast differentiation
Abstract
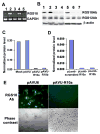
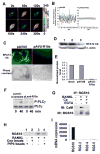
Significant progress has been made in studies of the mechanisms by which RANKL induces terminal osteoclast differentiation. However, many crucial details in the RANKL-evoked signaling pathway for osteoclast differentiation remain to be defined. We characterized genes specifically expressed in osteoclasts by differential screening of a human osteoclastoma cDNA library, and found that the regulator of G-protein signaling 10A (RGS10A), but not the RGS10B isoform, was specifically expressed in human osteoclasts. The expression of RGS10A is also induced by RANKL in osteoclast precursors and is prominently expressed in mouse osteoclast-like cells. RGS10A silencing by RNA interference blocked intracellular [Ca2+]i oscillations, the expression of NFAT2, and osteoclast terminal differentiation in both bone marrow cells and osteoclast precursor cell lines. Reintroduction of RGS10A rescued the impaired osteoclast differentiation. RGS10A silencing also resulted in premature osteoclast apoptosis. RGS10A silencing affected the RANKL-[Ca2+]i oscillation-NFAT2 signaling pathway but not other RANKL-induced responses. Our data demonstrate that target components of RGS10A are distinct from those of RGS12 in the RANKL signaling mechanism. Our results thus show the specificity of RGS10A as a key component in the RANKL-evoked signaling pathway for osteoclast differentiation, which may present a promising target for therapeutic intervention.
Figures



Similar articles
-
RGS12 is essential for RANKL-evoked signaling for terminal differentiation of osteoclasts in vitro.J Bone Miner Res. 2007 Jan;22(1):45-54. doi: 10.1359/jbmr.061007. J Bone Miner Res. 2007. PMID: 17042716 Free PMC article.
-
RGS10-null mutation impairs osteoclast differentiation resulting from the loss of [Ca2+]i oscillation regulation.Genes Dev. 2007 Jul 15;21(14):1803-16. doi: 10.1101/gad.1544107. Epub 2007 Jul 12. Genes Dev. 2007. PMID: 17626792 Free PMC article.
-
Upstream stimulatory factors regulate OSCAR gene expression in RANKL-mediated osteoclast differentiation.J Mol Biol. 2008 Nov 14;383(3):502-11. doi: 10.1016/j.jmb.2008.08.036. Epub 2008 Aug 22. J Mol Biol. 2008. PMID: 18762192
-
[RANKL signaling and bone diseases. Quiescent osteoclast precursors and RANKL signaling].Clin Calcium. 2011 Aug;21(8):1187-92. Clin Calcium. 2011. PMID: 21814024 Review. Japanese.
-
New regulation mechanisms of osteoclast differentiation.Ann N Y Acad Sci. 2011 Dec;1240:E13-8. doi: 10.1111/j.1749-6632.2011.06373.x. Ann N Y Acad Sci. 2011. PMID: 22360322 Review.
Cited by
-
Atp6v1c1 may regulate filament actin arrangement in breast cancer cells.PLoS One. 2014 Jan 15;9(1):e84833. doi: 10.1371/journal.pone.0084833. eCollection 2014. PLoS One. 2014. PMID: 24454753 Free PMC article.
-
Regulator of G protein signaling 10: Structure, expression and functions in cellular physiology and diseases.Cell Signal. 2020 Nov;75:109765. doi: 10.1016/j.cellsig.2020.109765. Epub 2020 Aug 31. Cell Signal. 2020. PMID: 32882407 Free PMC article. Review.
-
Targeting Atp6v1c1 Prevents Inflammation and Bone Erosion Caused by Periodontitis and Reveals Its Critical Function in Osteoimmunology.PLoS One. 2015 Aug 14;10(8):e0134903. doi: 10.1371/journal.pone.0134903. eCollection 2015. PLoS One. 2015. PMID: 26274612 Free PMC article.
-
Role of regulator of G protein signaling proteins in bone.Front Biosci (Landmark Ed). 2014 Jan 1;19(4):634-48. doi: 10.2741/4232. Front Biosci (Landmark Ed). 2014. PMID: 24389209 Free PMC article. Review.
-
Inhibition of Rgs10 Expression Prevents Immune Cell Infiltration in Bacteria-induced Inflammatory Lesions and Osteoclast-mediated Bone Destruction.Bone Res. 2013 Sep 1;1(3):267-281. doi: 10.4248/BR201303005. Bone Res. 2013. PMID: 24761229 Free PMC article.
References
-
- Alliston T, Derynck R. Medicine: interfering with bone remodelling. Nature. 2002;416:686–687. - PubMed
-
- Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation 1. Nature. 2003;423:337–342. - PubMed
-
- Chambers TJ, Fuller K, Darby JA, Pringle JA, Horton MA. Monoclonal antibodies against osteoclasts inhibit bone resorption in vitro. Bone Miner. 1986;1:127–135. - PubMed
-
- Chen W, Li YP. Generation of mouse osteoclastogenic cell lines immortalized with SV40 large T antigen. J Bone Miner Res. 1998;13:1112–1123. - PubMed
-
- Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109:S67–S79. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

