Distinct sequence elements of cyclin B1 promote localization to chromatin, centrosomes, and kinetochores during mitosis
- PMID: 17881737
- PMCID: PMC2096604
- DOI: 10.1091/mbc.e06-06-0539
Distinct sequence elements of cyclin B1 promote localization to chromatin, centrosomes, and kinetochores during mitosis
Abstract
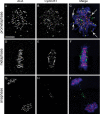
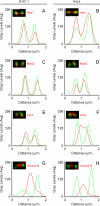
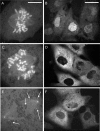
The mitotic cyclins promote cell division by binding and activating cyclin-dependent kinases (CDKs). Each cyclin has a unique pattern of subcellular localization that plays a vital role in regulating cell division. During mitosis, cyclin B1 is known to localize to centrosomes, microtubules, and chromatin. To determine the mechanisms of cyclin B1 localization in M phase, we imaged full-length and mutant versions of human cyclin B1-enhanced green fluorescent protein in live cells by using spinning disk confocal microscopy. In addition to centrosome, microtubule, and chromatin localization, we found that cyclin B1 also localizes to unattached kinetochores after nuclear envelope breakdown. Kinetochore recruitment of cyclin B1 required the kinetochore proteins Hec1 and Mad2, and it was stimulated by microtubule destabilization. Mutagenesis studies revealed that cyclin B1 is recruited to kinetochores through both CDK1-dependent and -independent mechanisms. In contrast, localization of cyclin B1 to chromatin and centrosomes is independent of CDK1 binding. The N-terminal domain of cyclin B1 is necessary and sufficient for chromatin association, whereas centrosome recruitment relies on sequences within the cyclin box. Our data support a role for cyclin B1 function at unattached kinetochores, and they demonstrate that separable and distinct sequence elements target cyclin B1 to kinetochores, chromatin, and centrosomes during mitosis.
Figures






References
-
- Acquaviva C., Herzog F., Kraft C., Pines J. The anaphase promoting complex/cyclosome is recruited to centromeres by the spindle assembly checkpoint. Nat. Cell Biol. 2004;6:892–898. - PubMed
-
- Arnaoutov A., Dasso M. The Ran GTPase regulates kinetochore function. Dev. Cell. 2003;5:99–111. - PubMed
-
- Bailly E., Cabantous S., Sondaz D., Bernadac A., Simon M. N. Differential cellular localization among mitotic cyclins from Saccharomyces cerevisiae: a new role for the axial budding protein Bud3 in targeting Clb2 to the mother-bud neck. J. Cell Sci. 2003;116:4119–4130. - PubMed
-
- Brown N. R., Lowe E. D., Petri E., Skamnaki V., Antrobus R., Johnson L. N. Cyclin B and cyclin a confer different substrate recognition properties on CDK2. Cell Cycle. 2007;6:1350–1359. - PubMed
-
- Brown N. R., Noble M. E., Endicott J. A., Johnson L. N. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1999;1:438–443. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

