The Edinburgh human metabolic network reconstruction and its functional analysis
- PMID: 17882155
- PMCID: PMC2013923
- DOI: 10.1038/msb4100177
The Edinburgh human metabolic network reconstruction and its functional analysis
Abstract
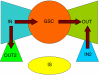
A better understanding of human metabolism and its relationship with diseases is an important task in human systems biology studies. In this paper, we present a high-quality human metabolic network manually reconstructed by integrating genome annotation information from different databases and metabolic reaction information from literature. The network contains nearly 3000 metabolic reactions, which were reorganized into about 70 human-specific metabolic pathways according to their functional relationships. By analysis of the functional connectivity of the metabolites in the network, the bow-tie structure, which was found previously by structure analysis, is reconfirmed. Furthermore, the distribution of the disease related genes in the network suggests that the IN (substrates) subset of the bow-tie structure has more flexibility than other parts.
Figures


References
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 - PMC - PubMed
-
- Csete M, Doyle J (2004) Bow ties, metabolism and disease. Trends Biotechnol 22: 446–450 - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

