Lipids as modulators of membrane fusion mediated by viral fusion proteins
- PMID: 17882414
- PMCID: PMC7080115
- DOI: 10.1007/s00249-007-0201-z
Lipids as modulators of membrane fusion mediated by viral fusion proteins
Abstract
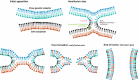
Enveloped viruses infect host cells by fusion of viral and target membranes. This fusion event is triggered by specific glycoproteins in the viral envelope. Fusion glycoproteins belong to either class I, class II or the newly described third class, depending upon their arrangement at the surface of the virion, their tri-dimensional structure and the location within the protein of a short stretch of hydrophobic amino acids called the fusion peptide, which is able to induce the initial lipid destabilization at the onset of fusion. Viral fusion occurs either with the plasma membrane for pH-independent viruses, or with the endosomal membranes for pH-dependent viruses. Although, viral fusion proteins are parted in three classes and the subcellular localization of fusion might vary, these proteins have to act, in common, on lipid assemblies. Lipids contribute to fusion through their physical, mechanical and/or chemical properties. Lipids can thus play a role as chemically defined entities, or through their preferential partitioning into membrane microdomains called "rafts", or by modulating the curvature of the membranes involved in the fusion process. The purpose of this review is to make a state of the art on recent findings on the contribution of cholesterol, sphingolipids and glycolipids in cell entry and membrane fusion of a number of viral families, whose members bear either class I or class II fusion proteins, or fusion proteins of the recently discovered third class.
Figures



References
-
- Alazard-Dany N, Ottmann Terrangle M, Volchkov V. Ebola and Marburg viruses: the humans strike back. Med Sci (Paris) 2006;22:405–410. - PubMed
-
- Alfsen A, Bomsel M. HIV-1 gp41 envelope residues 650-685 exposed on native virus act as a lectin to bind epithelial cell galactosyl ceramide. J Biol Chem. 2002;277:25649–25659. - PubMed
-
- Aman MJ, Bosio CM, Panchal RG, Burnett JC, Schmaljohn A, Bavari S. Molecular mechanisms of filovirus cellular trafficking. Microbes Infect. 2003;5:639–649. - PubMed
-
- Bartosch B, Cosset FL. Cell entry of hepatitis C virus. Virology. 2006;348:1–12. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

