Relevance of BAC transgene copy number in mice: transgene copy number variation across multiple transgenic lines and correlations with transgene integrity and expression
- PMID: 17882484
- PMCID: PMC3110064
- DOI: 10.1007/s00335-007-9056-y
Relevance of BAC transgene copy number in mice: transgene copy number variation across multiple transgenic lines and correlations with transgene integrity and expression
Abstract
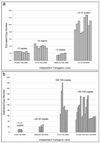
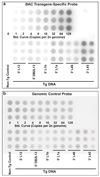
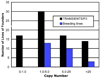
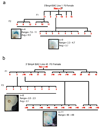
Bacterial artificial chromosomes (BACs) are excellent tools for manipulating large DNA fragments and, as a result, are increasingly utilized to engineer transgenic mice by pronuclear injection. The demand for BAC transgenic mice underscores the need for careful inspection of BAC integrity and fidelity following transgenesis, which may be crucial for interpreting transgene function. Thus, it is imperative that reliable methods for assessing these parameters are available. However, there are limited data regarding whether BAC transgenes routinely integrate in the mouse genome as intact molecules, how BAC transgenes behave as they are passed through the germline across successive generations, and how variation in BAC transgene copy number relates to transgene expression. To address these questions, we used TaqMan real-time PCR to estimate BAC transgene copy number in BAC transgenic embryos and lines. Here we demonstrate the reproducibility of copy number quantification with this method and describe the variation in copy number across independent transgenic lines. In addition, polymorphic marker analysis suggests that the majority of BAC transgenic lines contain intact molecules. Notably, all lines containing multiple BAC copies also contain all BAC-specific markers. Three of 23 founders analyzed contained BAC transgenes integrated into more than one genomic location. Finally, we show increased BAC transgene copy number correlates with increased BAC transgene expression. In sum, our efforts have provided a reliable method for assaying BAC transgene integrity and fidelity, and data that should be useful for researchers using BACs as transgenic vectors.
Figures




References
-
- Alexander GM, Erwin KL, Byers N, Deitch JS, Augelli BJ, et al. Effect of transgene copy number on survival in the G93A SOD1 transgenic mouse model of ALS. Brain Res Mol Brain Res. 2004;130:7–15. - PubMed
-
- Ballester M, Castello A, Ibanez E, Sanchez A, Folch JM. Real-time quantitative PCR-based system for determining transgene copy number in transgenic animals. Biotechniques. 2004;37:610–613. - PubMed
-
- Bishop J. Chromosomal insertion of foreign DNA. Reprod Nutr Dev. 1996;36:607–618. - PubMed
-
- Bishop JO, Smith P. Mechanism of chromosomal integration of microinjected DNA. Mol Biol Med. 1989;6:283–298. - PubMed
-
- Bitgood MJ, McMahon AP. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev Biol. 1995;172:126–138. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

