Factors affecting guanine nucleotide binding to rat AMPA receptors
- PMID: 17884024
- PMCID: PMC2078237
- DOI: 10.1016/j.brainres.2007.08.021
Factors affecting guanine nucleotide binding to rat AMPA receptors
Abstract
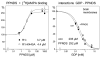
Glutamate receptors are competitively inhibited by guanine nucleotides. Insight into the physiological function of this inhibition would be greatly advanced if nucleotide binding could be eliminated through mutations without altering other aspects of receptor function, or if compounds were discovered that selectively prevent nucleotide binding. It was previously reported that a lysine in the chick kainate binding protein (cKBP) is specifically involved in guanine nucleotide binding. In the present study we mutated the equivalent lysine in the rat AMPA receptor subunit GluR1 flip to alanine (K445A) and assessed changes in nucleotide affinity from the displacement of [(3)H]fluorowillardiine. As in the cKBP, the affinity for nucleotides was greatly reduced while the binding affinity for agonists remained unchanged. The reduction in affinity was largest for GTP (factor of 5.8) and GDP (4.4) and minor for GMP and guanosine. This suggests that K445 is involved in stabilizing the second phosphate of the nucleotide. Given that bulkier analogs like GDP-fucose are also accommodated at this site, it seems likely that nucleotides bind in such a way that their phosphates project out of the cleft. In excised-patch recordings using short pulses of glutamate, the K445A mutation increased the EC(50) for the peak response 1.8-fold and accelerated desensitization and deactivation. This indicates that the effects of this mutation are not as specific as previously suggested. Efforts to selectively eliminate inhibition by nucleotides may therefore depend on mapping out further the docking site. In a first attempt using point mutations we ruled out several amino acids around the cleft as being involved in nucleotide binding. Also, the AMPA receptor modulator PPNDS which competitively inhibits nucleotide binding to purinergic receptors did not affect nucleotide inhibition, suggesting that there are major differences in the topography between purinergic and glutamate receptors. Thus new approaches, including crystallography, may be called for to identify residues uniquely involved in nucleotide binding.
Figures

Similar articles
-
Characterisation of the interaction between guanyl nucleotides and AMPA receptors in rat brain.Neuropharmacology. 1996;35(11):1583-93. doi: 10.1016/s0028-3908(96)00123-2. Neuropharmacology. 1996. PMID: 9025106
-
Effects of thiocyanate and AMPA receptor ligands on (S)-5-fluorowillardiine, (S)-AMPA and (R,S)-AMPA binding.Eur J Pharmacol. 1997 Jun 25;329(2-3):213-21. Eur J Pharmacol. 1997. PMID: 9226415
-
Divergent effects of the purinoceptor antagonists suramin and pyridoxal-5'-phosphate-6-(2'-naphthylazo-6'-nitro-4',8'-disulfonate) (PPNDS) on alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors.Mol Pharmacol. 2004 Dec;66(6):1738-47. doi: 10.1124/mol.104.003038. Epub 2004 Sep 24. Mol Pharmacol. 2004. PMID: 15448189
-
Regulation of agonist binding to A2A adenosine receptors: effects of guanine nucleotides (GDP[S] and GTP[S]) and Mg2+ ion.Biochim Biophys Acta. 1993 Dec 16;1220(1):76-84. doi: 10.1016/0167-4889(93)90100-4. Biochim Biophys Acta. 1993. PMID: 8268248
-
Receptor activation of G proteins.FASEB J. 1988 Oct;2(13):2841-8. doi: 10.1096/fasebj.2.13.3139484. FASEB J. 1988. PMID: 3139484 Review.
References
-
- Arai A, Kessler M, Ambros-Ingerson J, Quan A, Yigiter E, Rogers G, Lynch G. Effects of a centrally active benzoylpyrrolidine drug on AMPA receptor kinetics. Neuroscience. 1996;75:573–585. - PubMed
-
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. - PubMed
-
- Baron BM, Dudley MW, McCarty DR, Miller FP, Reynolds IJ, Schmidt CJ. Guanine nucleotides are competitive inhibitors of N-methyl-D-aspartate at its receptor site both in vitro and in vivo. J Pharmacol Exp Ther. 1989;250:162–169. - PubMed
-
- Bellamacina CR. The nicotinamide dinucleotide binding motif: a comparison of nucleotide binding proteins. FASEB J. 1996;10:1257–1269. - PubMed
-
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principles of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

