A phenotypic small-molecule screen identifies an orphan ligand-receptor pair that regulates neural stem cell differentiation
- PMID: 17884634
- PMCID: PMC2758915
- DOI: 10.1016/j.chembiol.2007.07.016
A phenotypic small-molecule screen identifies an orphan ligand-receptor pair that regulates neural stem cell differentiation
Abstract
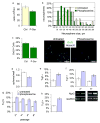
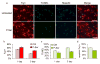
High-throughput identification of small molecules that selectively modulate molecular, cellular, or systems-level properties of the mammalian brain is a significant challenge. Here we report the chemical genetic identification of the orphan ligand phosphoserine (P-Ser) as an enhancer of neurogenesis. P-Ser inhibits neural stem cell/progenitor proliferation and self-renewal, enhances neurogenic fate commitment, and improves neuronal survival. We further demonstrate that the effects of P-Ser are mediated by the group III metabotropic glutamate receptor 4 (mGluR4). siRNA-mediated knockdown of mGluR4 abolished the effects of P-Ser and increased neurosphere proliferation, at least in part through upregulation of mTOR pathway activity. We also found that P-Ser increases neurogenesis in human embryonic stem cell-derived neural progenitors. This work highlights the tremendous potential of developing effective small-molecule drugs for use in regenerative medicine or transplantation therapy.
Figures




Comment in
-
An "orphan" finds a home in NSC regulation.Chem Biol. 2007 Sep;14(9):974-5. doi: 10.1016/j.chembiol.2007.09.001. Chem Biol. 2007. PMID: 17884628
Similar articles
-
An "orphan" finds a home in NSC regulation.Chem Biol. 2007 Sep;14(9):974-5. doi: 10.1016/j.chembiol.2007.09.001. Chem Biol. 2007. PMID: 17884628
-
Activation of metabotropic glutamate receptor 4 regulates proliferation and neural differentiation in neural stem/progenitor cells of the rat subventricular zone and increases phosphatase and tensin homolog protein expression.J Neurochem. 2021 Feb;156(4):465-480. doi: 10.1111/jnc.14984. Epub 2020 Mar 12. J Neurochem. 2021. PMID: 32052426
-
Phosphoserine phosphatase is expressed in the neural stem cell niche and regulates neural stem and progenitor cell proliferation.Stem Cells. 2007 Aug;25(8):1975-84. doi: 10.1634/stemcells.2007-0046. Epub 2007 May 10. Stem Cells. 2007. PMID: 17495110
-
Cannabinoid receptor signaling in progenitor/stem cell proliferation and differentiation.Prog Lipid Res. 2013 Oct;52(4):633-50. doi: 10.1016/j.plipres.2013.05.004. Epub 2013 Sep 25. Prog Lipid Res. 2013. PMID: 24076098 Review.
-
[Functional glutamate signaling in neural progenitor cells].Yakugaku Zasshi. 2011;131(9):1311-6. doi: 10.1248/yakushi.131.1311. Yakugaku Zasshi. 2011. PMID: 21881304 Review. Japanese.
Cited by
-
Stem cells in drug screening for neurodegenerative disease.Korean J Physiol Pharmacol. 2012 Feb;16(1):1-9. doi: 10.4196/kjpp.2012.16.1.1. Epub 2012 Feb 28. Korean J Physiol Pharmacol. 2012. PMID: 22416213 Free PMC article.
-
Embryonic stem cell application in drug discovery.Acta Pharmacol Sin. 2011 Feb;32(2):152-9. doi: 10.1038/aps.2010.194. Epub 2011 Jan 10. Acta Pharmacol Sin. 2011. PMID: 21217770 Free PMC article. Review.
-
Multi-step usage of in vivo models during rational drug design and discovery.Int J Mol Sci. 2011;12(4):2262-74. doi: 10.3390/ijms12042262. Epub 2011 Apr 1. Int J Mol Sci. 2011. PMID: 21731440 Free PMC article. Review.
-
A High-content screen identifies compounds promoting the neuronal differentiation and the midbrain dopamine neuron specification of human neural progenitor cells.Sci Rep. 2015 Nov 6;5:16237. doi: 10.1038/srep16237. Sci Rep. 2015. PMID: 26542303 Free PMC article.
-
A novel balanced chromosomal translocation found in subjects with schizophrenia and schizotypal personality disorder: altered l-serine level associated with disruption of PSAT1 gene expression.Neurosci Res. 2011 Feb;69(2):154-60. doi: 10.1016/j.neures.2010.10.003. Epub 2010 Oct 16. Neurosci Res. 2011. PMID: 20955740 Free PMC article.
References
-
- Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, Rueger MA, Bae SK, Kittappa R, McKay RD. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. - PubMed
-
- Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, Stahl N, Yancopoulos GD, Greenberg ME. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–483. - PubMed
-
- Brazel CY, Nunez JL, Yang Z, Levison SW. Glutamate enhances survival and proliferation of neural progenitors derived from the subventricular zone. Neuroscience. 2005;131:55–65. - PubMed
-
- Canudas AM, Di Giorgi-Gerevini V, Iacovelli L, Nano G, D'Onofrio M, Arcella A, Giangaspero F, Busceti C, Ricci-Vitiani L, Battaglia G, et al. PHCCC, a specific enhancer of type 4 metabotropic glutamate receptors, reduces proliferation and promotes differentiation of cerebellar granule cell neuroprecursors. J Neurosci. 2004;24:10343–10352. - PMC - PubMed
-
- Capela A, Temple S. LeX/ssea-1 is expressed by adult mouse CNS stem cells, identifying them as nonependymal. Neuron. 2002;35:865–875. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

