The mitochondrial transcription termination factor mTERF modulates replication pausing in human mitochondrial DNA
- PMID: 17884915
- PMCID: PMC2095818
- DOI: 10.1093/nar/gkm676
The mitochondrial transcription termination factor mTERF modulates replication pausing in human mitochondrial DNA
Abstract
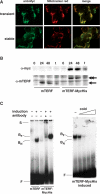
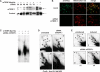
The mammalian mitochondrial transcription termination factor mTERF binds with high affinity to a site within the tRNA(Leu(UUR)) gene and regulates the amount of read through transcription from the ribosomal DNA into the remaining genes of the major coding strand of mitochondrial DNA (mtDNA). Electrophoretic mobility shift assays (EMSA) and SELEX, using mitochondrial protein extracts from cells induced to overexpress mTERF, revealed novel, weaker mTERF-binding sites, clustered in several regions of mtDNA, notably in the major non-coding region (NCR). Such binding in vivo was supported by mtDNA immunoprecipitation. Two-dimensional neutral agarose gel electrophoresis (2DNAGE) and 5' end mapping by ligation-mediated PCR (LM-PCR) identified the region of the canonical mTERF-binding site as a replication pause site. The strength of pausing was modulated by the expression level of mTERF. mTERF overexpression also affected replication pausing in other regions of the genome in which mTERF binding was found. These results indicate a role for TERF in mtDNA replication, in addition to its role in transcription. We suggest that mTERF could provide a system for coordinating the passage of replication and transcription complexes, analogous with replication pause-region binding proteins in other systems, whose main role is to safeguard the integrity of the genome whilst facilitating its efficient expression.
Figures




References
-
- Asin-Cayuela J, Gustafsson CM. Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem. Sci. 2007;32:111–117. - PubMed
-
- Fernandez-Silva P, Enriquez JA, Montoya J. Replication and transcription of mammalian mitochondrial DNA. Exp. Physiol. 2003;88:41–56. - PubMed
-
- Kruse B, Narasimhan N, Attardi G. Termination of transcription in human mitochondria: identification and purification of a DNA-binding protein factor that promotes termination. Cell. 1989;58:391–397. - PubMed
-
- Daga A, Micol V, Hess D, Aebersold R, Attardi G. Molecular characterization of the transcription termination factor from human mitochondria. J. Biol. Chem. 1993;268:8123–8130. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

